(-)-Kusunokinin as a Potential Aldose Reductase Inhibitor: Equivalency Observed via AKR1B1 Dynamics Simulation
- PMID: 33458512
- PMCID: PMC7807751
- DOI: 10.1021/acsomega.0c05102
(-)-Kusunokinin as a Potential Aldose Reductase Inhibitor: Equivalency Observed via AKR1B1 Dynamics Simulation
Abstract
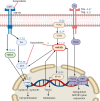
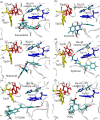
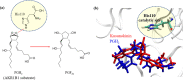
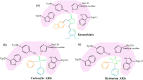
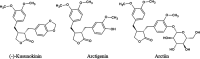
(-)-Kusunokinin performed its anticancer potency through CFS1R and AKT pathways. Its ambiguous binding target has, however, hindered the next development phase. Our study thus applied molecular docking and molecular dynamics simulation to predict the protein target from the pathways. Among various candidates, aldo-keto reductase family 1 member B1 (AKR1B1) was finally identified as a (-)-kusunokinin receptor. The predicted binding affinity of (-)-kusunokinin was better than the selected aldose reductase inhibitors (ARIs) and substrates. The compound also had no significant effect on AKR1B1 conformation. An intriguing AKR1B1 efficacy, with respect to the known inhibitors (epalrestat, zenarestat, and minalrestat) and substrates (UVI2008 and prostaglandin H2), as well as a similar interactive insight of the enzyme pocket, pinpointed an ARI equivalence of (-)-kusunokinin. An aromatic ring and a γ-butyrolactone ring shared a role with structural counterparts in known inhibitors. The modeling explained that the aromatic constituent contributed to π-π attraction with Trp111. In addition, the γ-butyrolactone ring bound the catalytic His110 using hydrogen bonds, which could lead to enzymatic inhibition as a consequence of substrate competitiveness. Our computer-based findings suggested that the potential of (-)-kusunokinin could be furthered by in vitro and/or in vivo experiments to consolidate (-)-kusunokinin as a new AKR1B1 antagonist in the future.
© 2020 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Trans-(±)-Kusunokinin Binding to AKR1B1 Inhibits Oxidative Stress and Proteins Involved in Migration in Aggressive Breast Cancer.Antioxidants (Basel). 2022 Nov 27;11(12):2347. doi: 10.3390/antiox11122347. Antioxidants (Basel). 2022. PMID: 36552555 Free PMC article.
-
Exploring the interactional details between aldose reductase (AKR1B1) and 3-Mercapto-5H-1,2,4-triazino[5,6-b]indole-5-acetic acid through molecular dynamics simulations.J Biomol Struct Dyn. 2019 Apr;37(7):1724-1735. doi: 10.1080/07391102.2018.1465851. Epub 2018 May 30. J Biomol Struct Dyn. 2019. PMID: 29671687
-
Potential Stereoselective Binding of Trans-(±)-Kusunokinin and Cis-(±)-Kusunokinin Isomers to CSF1R.Molecules. 2022 Jun 29;27(13):4194. doi: 10.3390/molecules27134194. Molecules. 2022. PMID: 35807438 Free PMC article.
-
Role of aldo-keto reductase family 1 member B1 (AKR1B1) in the cancer process and its therapeutic potential.J Cell Mol Med. 2020 Aug;24(16):8890-8902. doi: 10.1111/jcmm.15581. Epub 2020 Jul 6. J Cell Mol Med. 2020. PMID: 32633024 Free PMC article. Review.
-
Aldo-keto reductase family 1 member B1 inhibitors: old drugs with new perspectives.Recent Pat Anticancer Drug Discov. 2009 Nov;4(3):246-53. doi: 10.2174/157489209789206931. Recent Pat Anticancer Drug Discov. 2009. PMID: 19522700 Review.
Cited by
-
Phyto-Computational Intervention of Diabetes Mellitus at Multiple Stages Using Isoeugenol from Ocimum tenuiflorum: A Combination of Pharmacokinetics and Molecular Modelling Approaches.Molecules. 2022 Sep 22;27(19):6222. doi: 10.3390/molecules27196222. Molecules. 2022. PMID: 36234759 Free PMC article.
-
Trans-(-)-Kusunokinin: A Potential Anticancer Lignan Compound against HER2 in Breast Cancer Cell Lines?Molecules. 2021 Jul 27;26(15):4537. doi: 10.3390/molecules26154537. Molecules. 2021. PMID: 34361688 Free PMC article.
-
Trans-(±)-TTPG-B Attenuates Cell Cycle Progression and Inhibits Cell Proliferation on Cholangiocarcinoma Cells.Molecules. 2023 Oct 30;28(21):7342. doi: 10.3390/molecules28217342. Molecules. 2023. PMID: 37959760 Free PMC article.
-
Trans-(±)-Kusunokinin Binding to AKR1B1 Inhibits Oxidative Stress and Proteins Involved in Migration in Aggressive Breast Cancer.Antioxidants (Basel). 2022 Nov 27;11(12):2347. doi: 10.3390/antiox11122347. Antioxidants (Basel). 2022. PMID: 36552555 Free PMC article.
-
In Silico Identification of Potential Sites for a Plastic-Degrading Enzyme by a Reverse Screening through the Protein Sequence Space and Molecular Dynamics Simulations.Molecules. 2022 May 23;27(10):3353. doi: 10.3390/molecules27103353. Molecules. 2022. PMID: 35630830 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

