Characterization and Application of Precore/Core-Related Antigens in Animal Models of Hepatitis B Virus Infection
- PMID: 33458844
- PMCID: PMC8286267
- DOI: 10.1002/hep.31720
Characterization and Application of Precore/Core-Related Antigens in Animal Models of Hepatitis B Virus Infection
Erratum in
-
Erratum.Hepatology. 2022 Aug;76(2):533. doi: 10.1002/hep.32507. Epub 2022 Apr 22. Hepatology. 2022. PMID: 35451512 No abstract available.
Abstract
Background and aims: The hepatitis B core-related antigen (HBcrAg), a composite antigen of precore/core gene including classical hepatitis B core protein (HBc) and HBeAg and, additionally, the precore-related antigen PreC, retaining the N-terminal signal peptide, has emerged as a surrogate marker to monitor the intrahepatic HBV covalently closed circular DNA (cccDNA) and to define meaningful treatment endpoints.
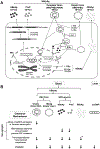
Approach and results: Here, we found that the woodchuck hepatitis virus (WHV) precore/core gene products (i.e., WHV core-related antigen [WHcrAg]) include the WHV core protein and WHV e antigen (WHeAg) as well as the WHV PreC protein (WPreC) in infected woodchucks. Unlike in HBV infection, WHeAg and WPreC proteins were N-glycosylated, and no significant amounts of WHV empty virions were detected in WHV-infected woodchuck serum. WHeAg was the predominant form of WHcrAg, and a positive correlation was found between the serum WHeAg and intrahepatic cccDNA. Both WHeAg and WPreC antigens displayed heterogeneous proteolytic processing at their C-termini, resulting in multiple species. Analysis of the kinetics of each component of the precore/core-related antigen, along with serum viral DNA and surface antigens, in HBV-infected chimpanzees and WHV-infected woodchucks revealed multiple distinct phases of viral decline during natural resolution and in response to antiviral treatments. A positive correlation was found between HBc and intrahepatic cccDNA but not between HBeAg or HBcrAg and cccDNA in HBV-infected chimpanzees, suggesting that HBc can be a better marker for intrahepatic cccDNA.
Conclusions: In conclusion, careful monitoring of each component of HBcrAg along with other classical markers will help understand intrahepatic viral activities to elucidate natural resolution mechanisms as well as guide antiviral development.
© 2021 by the American Association for the Study of Liver Diseases.
Conflict of interest statement
Figures


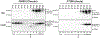
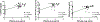


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

