Class III phosphatidylinositol 3-kinase complex I subunit NRBF2/Atg38 - from cell and structural biology to health and disease
- PMID: 33459128
- PMCID: PMC8726667
- DOI: 10.1080/15548627.2021.1872240
Class III phosphatidylinositol 3-kinase complex I subunit NRBF2/Atg38 - from cell and structural biology to health and disease
Abstract
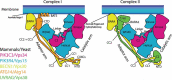
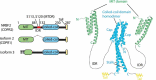
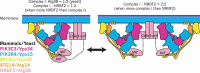
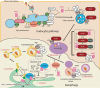
Macroautophagy/autophagy is triggered by various starvation and stress conditions. The phospholipid phosphatidylinositol-3-phosphate (PtdIns3P) is essential for the formation of the autophagosome both in yeast and mammals. The class III phosphatidylinositol 3-kinase, PIK3C3C in humans or Vps34 in yeast, produces PtdIns3P by phosphorylating the 3'-OH position of phosphatidylinositol (PtdIns). In order to synthesize PtdIns3P for the initiation of autophagy, PIK3C3/Vps34 has a heterotetrameric core, the PIK3C3 complex I (hereafter complex I) composed of PIK3C3/Vps34, PIK3R4/Vps15, BECN1/Vps30, and ATG14/Atg14. A fifth component of complex I, NRBF2 in mammals and Atg38 in yeast, was found and has been characterized in the past decade. The field has been expanding from cell and structural biology to mouse model and cohort studies. Here I will summarize the structures and models of complex I binding NRBF2/Atg38, its intracellular roles, and its involvement in health and disease. Along with this expansion of the field, different conclusions have been drawn in several topics. I will clarify what has and has not been agreed, and what is to be clarified in the future.
Keywords: atg38; membranes; nrbf2; pik3c3; ptdins3p; vps34.
Conflict of interest statement
The authors report no conflict of interest.
Figures

Similar articles
-
Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.Autophagy. 2016 Nov;12(11):2129-2144. doi: 10.1080/15548627.2016.1226736. Epub 2016 Sep 14. Autophagy. 2016. PMID: 27630019 Free PMC article.
-
MTORC1-mediated NRBF2 phosphorylation functions as a switch for the class III PtdIns3K and autophagy.Autophagy. 2017 Mar 4;13(3):592-607. doi: 10.1080/15548627.2016.1269988. Epub 2017 Jan 6. Autophagy. 2017. PMID: 28059666 Free PMC article.
-
Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.Autophagy. 2019 Aug;15(8):1333-1355. doi: 10.1080/15548627.2019.1581009. Epub 2019 Mar 4. Autophagy. 2019. PMID: 30767700 Free PMC article.
-
Activation Mechanisms of the VPS34 Complexes.Cells. 2021 Nov 11;10(11):3124. doi: 10.3390/cells10113124. Cells. 2021. PMID: 34831348 Free PMC article. Review.
-
WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome.J Cell Sci. 2015 Jan 15;128(2):207-17. doi: 10.1242/jcs.146258. J Cell Sci. 2015. PMID: 25568150 Review.
Cited by
-
Assessing the Presence of Phosphoinositides on Autophagosomal Membrane in Yeast by Live Cell Imaging.Microorganisms. 2024 Jul 18;12(7):1458. doi: 10.3390/microorganisms12071458. Microorganisms. 2024. PMID: 39065227 Free PMC article.
-
NRBF2 plays a crucial role in the acquisition process of learning and memory, independent of the Vps34 complex.Front Behav Neurosci. 2025 Feb 12;19:1529522. doi: 10.3389/fnbeh.2025.1529522. eCollection 2025. Front Behav Neurosci. 2025. PMID: 40013119 Free PMC article.
-
The Multifaceted Role of Autophagy in Nasopharyngeal Carcinoma: Translational Perspectives on Pathogenesis, Biomarkers, Treatment Resistance, and Emerging Therapies.Cancers (Basel). 2025 Aug 5;17(15):2577. doi: 10.3390/cancers17152577. Cancers (Basel). 2025. PMID: 40805271 Free PMC article. Review.
-
Autophagy in Disease Onset and Progression.Aging Dis. 2024 Aug 1;15(4):1646-1671. doi: 10.14336/AD.2023.0815. Aging Dis. 2024. PMID: 37962467 Free PMC article. Review.
-
The Role of Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 in the Pathogenesis of Human Cancer.Int J Mol Sci. 2021 Oct 11;22(20):10964. doi: 10.3390/ijms222010964. Int J Mol Sci. 2021. PMID: 34681622 Free PMC article. Review.
References
-
- Yasumo H, Masuda N, Furusawa T, et al. Nuclear receptor binding factor-2 (NRBF-2), a possible gene activator protein interacting with nuclear hormone receptors. Biochim Biophys Acta. 2000;1490(1–2):189–197. - PubMed
-
- Flores AM, Li L, Aneskievich BJ.. Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. J Invest Dermatol. 2004;123(6):1092–1101. - PubMed
-
- Schu PV, Takegawa K, Fry MJ, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260(5104):88–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
