Deletions in CWH43 cause idiopathic normal pressure hydrocephalus
- PMID: 33459505
- PMCID: PMC7933959
- DOI: 10.15252/emmm.202013249
Deletions in CWH43 cause idiopathic normal pressure hydrocephalus
Abstract
Idiopathic normal pressure hydrocephalus (iNPH) is a neurological disorder that occurs in about 1% of individuals over age 60 and is characterized by enlarged cerebral ventricles, gait difficulty, incontinence, and cognitive decline. The cause and pathophysiology of iNPH are largely unknown. We performed whole exome sequencing of DNA obtained from 53 unrelated iNPH patients. Two recurrent heterozygous loss of function deletions in CWH43 were observed in 15% of iNPH patients and were significantly enriched 6.6-fold and 2.7-fold, respectively, when compared to the general population. Cwh43 modifies the lipid anchor of glycosylphosphatidylinositol-anchored proteins. Mice heterozygous for CWH43 deletion appeared grossly normal but displayed hydrocephalus, gait and balance abnormalities, decreased numbers of ependymal cilia, and decreased localization of glycosylphosphatidylinositol-anchored proteins to the apical surfaces of choroid plexus and ependymal cells. Our findings provide novel mechanistic insights into the origins of iNPH and demonstrate that it represents a distinct disease entity.
Keywords: CWH43; GPI-anchored protein; hydrocephalus; normal pressure hydrocephalus.
© 2021 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Sanger DNA sequencing data from two iNPH patients confirming the presence of heterozygous CWH43 deletions. Arrows identify location of the deletion.
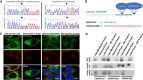
Diagram of the domain structure of the Cwh43 protein illustrating the location of the damaging CWH43 deletions. Lower panel details the disruptive effect of the k696Asnfs mutation on the C‐terminal endoplasmic reticulum export signal (YF), replacing it via a frameshift with a 23 amino acid sequence that contains an endoplasmic reticulum retention signal (KKKS).
Fluorescence micrographs of HeLa cells transfected with plasmids encoding human GFP‐Cwh43, GFP‐Cwh43‐k696fs, or GFP‐Cwh43‐Leu533ter fusion proteins. To label the ER, HeLa cells were transduced with a baculovirus encoding a fluorescently labeled RFP‐calreticulin fusion protein containing an ER retention signal (RFP‐calreticulin‐KDEL). To label the Golgi apparatus, cells were immunostained for golgin‐97 (red). When compared with wild‐type GFP‐Cwh43, GFP‐Cwh43‐k696fs, and GFP‐Cwh43‐Leu533 showed decreased association with the Golgi apparatus (but not the ER) and were diffusely distributed throughout the cytoplasm. Scale bar is approximately 5 µm.
Western blot analysis of total membrane, aqueous and lipid (GPI‐anchor‐containing) Triton X‐114 extracts derived from wild‐type HeLa cells, and two independent CRISPR CWH43 knockout (KO) HeLa cell lines in which a mutated CWH43 gene encodes a protein that is truncated near Leu533 and CWH43 mRNA and protein are markedly reduced. Cells were transfected to overexpress a control GFP plasmid, a plasmid encoding human wild‐type Cwh43 with GFP fused to the N‐terminus, or a plasmid encoding human CWH43 harboring the iNPH‐associated mutation (Lys696AsnfsTer23) with GFP fused to the N‐terminus. The Western blot was stained using an antibody directed against CD59, a GPI‐anchored protein.

mRNA in situ hybridization images showing expression of Cwh43 mRNA in the mouse brain. Enclosed areas containing potions of the ventricle, hippocampus, and dorsal thalamus are shown at higher magnification on the right. Arrowheads point to choroid plexus. Scale bar is approximately 400 µm.
Fluorescence immunohistochemistry of the ependymal surface of the lateral ventricle of the mouse brain. Cilia are visualized using an antibody for acetylated alpha tubulin (green). Cwh43 is visualized using a specific anti‐Cwh43 antibody (red). Nuclei are counterstained using DAPI (blue). Arrowheads point to motile cilia and scale bar is approximately 5 µm.
Confocal fluorescence immunocytochemistry images of a single cultured mouse ciliated ependymal cell. Cilia were visualized using an antibody for acetylated alpha tubulin (green). Cwh43 immunoreactivity was visualized using a specific anti‐Cwh43 antibody (red). Scale bar (left column) is approximately 4 µm. Images in the column on the right represent a Z‐stack reconstruction of confocal images showing localization of Cwh43 immunoreactivity in cilia of a mouse ependymal cell. Scale bar (right column) is approximately 5 µm.
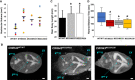
Scatter plot comparing ventricular volume from CWH43WT/WT (wild‐type, WT), heterozygous CWH43WT/M533, homozygous CWH43M533/M533, and CWH43M533/A530 mice at 6 months. Ventricular volume was calculated from T2‐weighted MR images of the mouse brain using a custom automated computer algorithm. Horizontal bars indicate the mean of the measurements in each column. Statistical significance for each mutant mouse line compared to WT was determined using the unpaired t‐test (P = 0.0015 for CWH43WT/M533; P = 0.0014 for CWH43M533/M533; P = 0.006 for CWH43M533/A530).
Representative 3D volumetric MR images of mouse brains from 6‐month‐old CWH43WT/WT, CWH43M533/M533, and CWH43M533/A530 mice. LV = lateral ventricle, 3rd V = third ventricle, 4th V = fourth ventricle. Scale bar is approximately 1 mm.
Quantitative gait analysis at 7 months of age revealed increased sway among homozygous CWH43M533/M533 mice, (*P = 0.03, n = 5) and compound heterozygous CWH43M533/A530 (*P = 0.034, n = 4) mice when compared to wild‐type CWH43WT/WT mice (n = 5). Sway (the distance between the hind paws during walking) was measured repeatedly for individual mice in each group during a constrained unidirectional walk. Data shown are the mean ± SD for each group. Statistical significance was determined using the unpaired t‐test.
Box plot showing rotarod performance data for CWH43WT/WT, CWH43WT/M533, CWH43M533/M533, and CWH43M533/A530 mice at 7 months of age. Data shown are the mean, 1st quartile, 3rd quartile, minimum, and maximum for each group of mice. Statistical significance was determined using the unpaired t‐test. When compared to wild‐type CWH43WT/WT mice (n = 10), balance time on the rotarod was decreased significantly among heterozygous CWH43WT/M533 mice (*P = 0.04, n = 8), homozygous CWH43M533/M533 mice (*P = 0.03, n = 7), and compound heterozygous CWH43M533/A530 mice (*P = 0.03, n = 4).
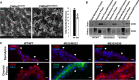
Scanning electron micrographs of the ependymal surface of the lateral ventricle of CWH43 WT and CWH43M533/M533 mice. Scale bar is approximately 40 µm. The graph on the right quantifies the data from the electron micrographs on the left. Data shown are the mean ± SEM. *P = 0.004, n = 4, unpaired t‐test.
Western blot analysis of total membrane, aqueous, and lipid (GPI‐anchor‐containing) Triton X‐114 extracts derived from wild‐type, CWH43M533/M533, and CWH43WT/M533 mouse brain or kidney. The Western blot was stained using an antibody directed against CD59, a GPI‐anchored protein.
Fluorescence immunohistochemistry for CD59 in the ependymal layer and choroid plexus of the lateral ventricle from CWH43WT/WT, CWH43M533/M533, and CWH43M533/A530 mice. Arrowheads point to apical surfaces of ependymal and choroid plexus cells. Nuclei are counterstained using DAPI (blue). Scale bar is approximately 5 µm.
Comment in
-
iNPH-the mystery resolving.EMBO Mol Med. 2021 Mar 5;13(3):e13720. doi: 10.15252/emmm.202013720. Epub 2021 Feb 8. EMBO Mol Med. 2021. PMID: 33555136 Free PMC article.
References
-
- Adams RD, Fisher CM, Hakim S, Ojemann RG, Sweet WH (1965) Symptomatic occult hydrocephalus with "Normal" cerebrospinal‐fluid pressure. A treatable syndrome. N Engl J Med 273: 117–126 - PubMed
-
- Allen Brain Atlas, authors. www.brain‐map.org/
-
- Del Bigio MR (1993) Neuropathological changes caused by hydrocephalus. Acta Neuropathol 85: 573–585 - PubMed
-
- Eide PK, Sorteberg W (2010) Diagnostic intracranial pressure monitoring and surgical management in idiopathic normal pressure hydrocephalus: a 6‐year review of 214 patients. Neurosurgery 66: 80–91 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

