SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights
- PMID: 33460172
- PMCID: PMC8013332
- DOI: 10.1002/jobm.202000537
SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights
Abstract
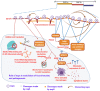
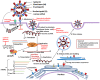
The outbreak of a novel coronavirus associated with acute respiratory disease, called COVID-19, marked the introduction of the third spillover of an animal coronavirus (CoV) to humans in the last two decades. The genome analysis with various bioinformatics tools revealed that the causative pathogen (SARS-CoV-2) belongs to the subgenus Sarbecovirus of the genus Betacoronavirus, with highly similar genome as bat coronavirus and receptor-binding domain (RBD) of spike glycoprotein as Malayan pangolin coronavirus. Based on its genetic proximity, SARS-CoV-2 is likely to have originated from bat-derived CoV and transmitted to humans via an unknown intermediate mammalian host, probably Malayan pangolin. Further, spike protein S1/S2 cleavage site of SARS-CoV-2 has acquired polybasic furin cleavage site which is absent in bat and pangolin suggesting natural selection either in an animal host before zoonotic transfer or in humans following zoonotic transfer. In the current review, we recapitulate a preliminary opinion about the disease, origin and life cycle of SARS-CoV-2, roles of virus proteins in pathogenesis, commonalities, and differences between different corona viruses. Moreover, the crystal structures of SARS-CoV-2 proteins with unique characteristics differentiating it from other CoVs are discussed. Our review also provides comprehensive information on the molecular aspects of SARS-CoV-2 including secondary structures in the genome and protein-protein interactions which can be useful to understand the aggressive spread of the SARS-CoV-2. The mutations and the haplotypes reported in the SARS-CoV-2 genome are summarized to understand the virus evolution.
Keywords: ACE2; SARS-CoV-2; angiotensin-converting enzyme 2; pandemic coronavirus; severe acute respiratory syndrome coronavirus 2.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures




References
-
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Gulyaeva AA, Haagmans BL, et al. The species and its viruses—a statement of the coronavirus study group. bioRxiv. 2020. 10.1101/2020.02.07.937862 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

