What coronavirus 3C-like protease tells us: From structure, substrate selectivity, to inhibitor design
- PMID: 33460213
- PMCID: PMC8014231
- DOI: 10.1002/med.21783
What coronavirus 3C-like protease tells us: From structure, substrate selectivity, to inhibitor design
Abstract
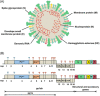
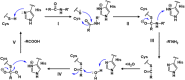
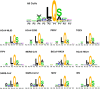
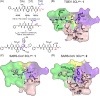
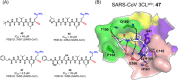
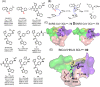
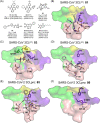
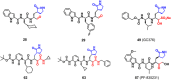
The emergence of a variety of coronaviruses (CoVs) in the last decades has posed huge threats to human health. Especially, the ongoing pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to more than 70 million infections and over 1.6 million of deaths worldwide in the past few months. None of the efficacious antiviral agents against human CoVs have been approved yet. 3C-like protease (3CLpro ) is an attractive target for antiviral intervention due to its essential role in processing polyproteins translated from viral RNA, and its conserved structural feature and substrate specificity among CoVs in spite of the sequence variation. This review focuses on all available crystal structures of 12 CoV 3CLpro s and their inhibitors, and intends to provide a comprehensive understanding of this protease from multiple aspects including its structural features, substrate specificity, inhibitor binding modes, and more importantly, to recapitulate the similarity and diversity among different CoV 3CLpro s and the structure-activity relationship of various types of inhibitors. Such an attempt could gain a deep insight into the inhibition mechanisms and drive future structure-based drug discovery targeting 3CLpro s.
Keywords: 3C-like protease; binding modes; coronavirus; inhibitors; structure and function.
© 2021 Wiley Periodicals LLC.
Figures










References
-
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121(1):190‐193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

