Development of prognostic signature based on RNA binding proteins related genes analysis in clear cell renal cell carcinoma
- PMID: 33461173
- PMCID: PMC7906138
- DOI: 10.18632/aging.202360
Development of prognostic signature based on RNA binding proteins related genes analysis in clear cell renal cell carcinoma
Abstract
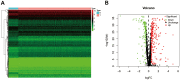
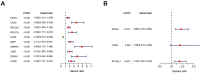
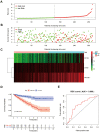
RNA binding proteins (RBPs) play significant roles in the development of tumors. However, a comprehensive analysis of the biological functions of RBPs in clear cell renal cell carcinoma (ccRCC) has not been performed. Our study aimed to construct an RBP-related risk model for prognosis prediction in ccRCC patients. First, RNA sequencing data of ccRCC were downloaded from The Cancer Genome Atlas (TCGA) database. Three RBP genes (EIF4A1, CARS, and RPL22L1) were validated as prognosis-related hub genes by univariate and multivariate Cox regression analyses and were integrated into a prognostic model by least absolute shrinkage and selection operator (LASSO) Cox regression analysis. According to this model, patients with high risk scores displayed significantly worse overall survival (OS) than those with low risk scores. Moreover, the multivariate Cox analysis results indicated that risk score, tumor grade, and tumor stage were significantly correlated with patient OS. A nomogram was constructed based on the three RBP genes and showed a good ability to predict outcomes in ccRCC patients. In conclusion, this study identified a three-RBP gene risk model for predicting the prognosis of patients, which is conducive to the identification of novel diagnostic and prognostic molecular markers.
Keywords: RNA binding proteins; biomarker; clear cell renal cell carcinoma; prognostic model.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

