The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities
- PMID: 33461542
- PMCID: PMC7812662
- DOI: 10.1186/s12943-020-01263-w
The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities
Abstract
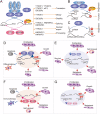
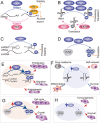
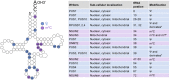
RNA modifications have recently emerged as critical posttranscriptional regulators of gene expression programmes. Significant advances have been made in understanding the functional role of RNA modifications in regulating coding and non-coding RNA processing and function, which in turn thoroughly shape distinct gene expression programmes. They affect diverse biological processes, and the correct deposition of many of these modifications is required for normal development. Alterations of their deposition are implicated in several diseases, including cancer. In this Review, we focus on the occurrence of N6-methyladenosine (m6A), 5-methylcytosine (m5C) and pseudouridine (Ψ) in coding and non-coding RNAs and describe their physiopathological role in cancer. We will highlight the latest insights into the mechanisms of how these posttranscriptional modifications influence tumour development, maintenance, and progression. Finally, we will summarize the latest advances on the development of small molecule inhibitors that target specific writers or erasers to rewind the epitranscriptome of a cancer cell and their therapeutic potential.
Keywords: 5-methylcytosine; Anti-cancer therapy; Cancer; Epitranscriptome; Inhibitors; Migration; N6-methyladenosine; Proliferation; Pseudouridine; RNA modifications; m5C; m6A; Ψ.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

