Functional analysis of tomato CHIP ubiquitin E3 ligase in heat tolerance
- PMID: 33462308
- PMCID: PMC7814054
- DOI: 10.1038/s41598-021-81372-8
Functional analysis of tomato CHIP ubiquitin E3 ligase in heat tolerance
Abstract
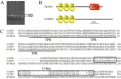
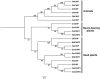
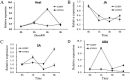
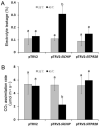
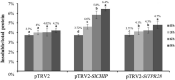
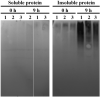
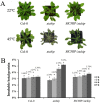
Plants have evolved genetic and physiological mechanisms to mitigate the adverse effects of high temperature. CARBOXYL TERMINUS OF THE HSC70-INTERACTING PROTEINS (CHIP) is a conserved chaperone-dependent ubiquitin E3 ligase that targets misfolded proteins. Here, we report functional analysis of the SlCHIP gene from tomato (Solanum lycopersicum) in heat tolerance. SlCHIP encodes a CHIP protein with three tandem tetracopeptide repeat (TPR) motifs and a C-terminal U box domain. Phylogenetic analysis of CHIP homologs from animals, spore-bearing and seed plants revealed a tree topology similar to the evolutionary tree of the organisms. Expression of SlCHIP was induced under high temperature and was also responsive to plant stress hormones. Silencing of SlCHIP in tomato reduced heat tolerance based on increased heat stress symptoms, reduced photosynthetic activity, elevated electrolyte leakage and accumulation of insoluble protein aggregates. The accumulated protein aggregates in SlCHIP-silenced plants were still highly ubiquitinated, suggesting involvement of other E3 ligases in ubiquitination. SlCHIP restored the heat tolerance of Arabidopsis chip mutant to the wild type levels. These results indicate that tomato SlCHIP plays a critical role in heat stress responses most likely by targeting degradation of misfolded proteins that are generated during heat stress.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The E3 ubiquitin ligase gene SlRING1 is essential for plant tolerance to cadmium stress in Solanum lycopersicum.J Biotechnol. 2020 Dec 20;324:239-247. doi: 10.1016/j.jbiotec.2020.11.008. Epub 2020 Nov 10. J Biotechnol. 2020. PMID: 33186659
-
Wheat CHIP E3 ubiquitin ligase, TaCEU, forms a complex with TaCHSP70 to regulate targeted proteins, enhancing thermotolerance in transgenic Arabidopsis.Plant Physiol Biochem. 2025 Apr;221:109615. doi: 10.1016/j.plaphy.2025.109615. Epub 2025 Feb 6. Plant Physiol Biochem. 2025. PMID: 39946907
-
E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses.PLoS Genet. 2014 Jan 30;10(1):e1004116. doi: 10.1371/journal.pgen.1004116. eCollection 2014 Jan. PLoS Genet. 2014. PMID: 24497840 Free PMC article.
-
Conserved and Unique Roles of Chaperone-Dependent E3 Ubiquitin Ligase CHIP in Plants.Front Plant Sci. 2021 Jul 9;12:699756. doi: 10.3389/fpls.2021.699756. eCollection 2021. Front Plant Sci. 2021. PMID: 34305988 Free PMC article. Review.
-
STUB1/CHIP: New insights in cancer and immunity.Biomed Pharmacother. 2023 Sep;165:115190. doi: 10.1016/j.biopha.2023.115190. Epub 2023 Jul 26. Biomed Pharmacother. 2023. PMID: 37506582 Review.
Cited by
-
Arabidopsis MED18 Interaction With RNA Pol IV and V Subunit NRPD2a in Transcriptional Regulation of Plant Immune Responses.Front Plant Sci. 2021 Oct 6;12:692036. doi: 10.3389/fpls.2021.692036. eCollection 2021. Front Plant Sci. 2021. PMID: 34691090 Free PMC article.
-
How Many Faces Does the Plant U-Box E3 Ligase Have?Int J Mol Sci. 2022 Feb 18;23(4):2285. doi: 10.3390/ijms23042285. Int J Mol Sci. 2022. PMID: 35216399 Free PMC article. Review.
-
Genome-wide profiling of histone (H3) lysine 4 (K4) tri-methylation (me3) under drought, heat, and combined stresses in switchgrass.BMC Genomics. 2024 Feb 29;25(1):223. doi: 10.1186/s12864-024-10068-w. BMC Genomics. 2024. PMID: 38424499 Free PMC article.
-
The ETHYLENE RESPONSE FACTOR6-GRETCHEN HAGEN3.5 module regulates rooting and heat tolerance in Dimocarpus longan.Plant Physiol. 2025 Mar 1;197(3):kiaf096. doi: 10.1093/plphys/kiaf096. Plant Physiol. 2025. PMID: 40106655 Free PMC article.
-
Crop Proteomics under Abiotic Stress: From Data to Insights.Plants (Basel). 2022 Oct 27;11(21):2877. doi: 10.3390/plants11212877. Plants (Basel). 2022. PMID: 36365330 Free PMC article. Review.
References
-
- Zhou S, Abaraha A. Response to heat stress in warm season and cool season turf grass cultivars. Sci. Res. Essays. 2007;2:95–100.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

