ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells
- PMID: 33462394
- PMCID: PMC7880902
- DOI: 10.1038/s41556-020-00624-3
ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells
Abstract
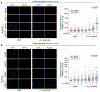
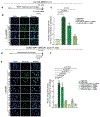
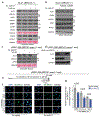
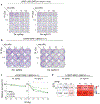
The response to poly(ADP-ribose) polymerase inhibitors (PARPi) is dictated by homologous recombination (HR) DNA repair and the abundance of lesions that trap PARP enzymes. It remains unclear, however, if the established role of PARP in promoting chromatin accessibility impacts viability in these settings. Using a CRISPR-based screen, we identified the PAR-binding chromatin remodeller ALC1/CHD1L as a key determinant of PARPi toxicity in HR-deficient cells. ALC1 loss reduced viability of breast cancer gene (BRCA)-mutant cells and enhanced sensitivity to PARPi by up to 250-fold, while overcoming several resistance mechanisms. ALC1 deficiency reduced chromatin accessibility concomitant with a decrease in the association of base damage repair factors. This resulted in an accumulation of replication-associated DNA damage, increased PARP trapping and a reliance on HR. These findings establish PAR-dependent chromatin remodelling as a mechanistically distinct aspect of PARPi responses and therapeutic target in HR-deficient cancers.
Conflict of interest statement
Competing interests
R.A.G. is a founder and scientific advisory board member of RADD Pharmaceuticals. No other authors declare any competing interests.
Figures













References
-
- Krishnakumar R et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

