Genetics of 35 blood and urine biomarkers in the UK Biobank
- PMID: 33462484
- PMCID: PMC7867639
- DOI: 10.1038/s41588-020-00757-z
Genetics of 35 blood and urine biomarkers in the UK Biobank
Erratum in
-
Author Correction: Genetics of 35 blood and urine biomarkers in the UK Biobank.Nat Genet. 2021 Nov;53(11):1622. doi: 10.1038/s41588-021-00956-2. Nat Genet. 2021. PMID: 34608296 Free PMC article. No abstract available.
Abstract
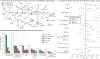
Clinical laboratory tests are a critical component of the continuum of care. We evaluate the genetic basis of 35 blood and urine laboratory measurements in the UK Biobank (n = 363,228 individuals). We identify 1,857 loci associated with at least one trait, containing 3,374 fine-mapped associations and additional sets of large-effect (>0.1 s.d.) protein-altering, human leukocyte antigen (HLA) and copy number variant (CNV) associations. Through Mendelian randomization (MR) analysis, we discover 51 causal relationships, including previously known agonistic effects of urate on gout and cystatin C on stroke. Finally, we develop polygenic risk scores (PRSs) for each biomarker and build 'multi-PRS' models for diseases using 35 PRSs simultaneously, which improved chronic kidney disease, type 2 diabetes, gout and alcoholic cirrhosis genetic risk stratification in an independent dataset (FinnGen; n = 135,500) relative to single-disease PRSs. Together, our results delineate the genetic basis of biomarkers and their causal influences on diseases and improve genetic risk stratification for common diseases.
Conflict of interest statement
Competing financial interests
The Board of Trustees of the Leland Stanford Junior University filed U.S. Provisional Application “Methods for diagnosis of polygenic diseases and phenotypes from genetic variation” (Serial No. 62/852,738) describing this work. J.K.P., M.A.R., N.S.-A., and Y.T. are designated as inventors of the patent. M.A.R is on the SAB of 54Gene and Computational Advisory Board for Goldfinch Bio and has advised BioMarin, Third Rock Ventures, MazeTx and Related Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Figures
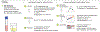



Comment in
-
Biomarker genetics.Nat Rev Nephrol. 2021 Apr;17(4):222. doi: 10.1038/s41581-021-00400-y. Nat Rev Nephrol. 2021. PMID: 33514940 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

