Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power
- PMID: 33462486
- PMCID: PMC7867648
- DOI: 10.1038/s41588-020-00766-y
Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power
Abstract
Admixed populations are routinely excluded from genomic studies due to concerns over population structure. Here, we present a statistical framework and software package, Tractor, to facilitate the inclusion of admixed individuals in association studies by leveraging local ancestry. We test Tractor with simulated and empirical two-way admixed African-European cohorts. Tractor generates accurate ancestry-specific effect-size estimates and P values, can boost genome-wide association study (GWAS) power and improves the resolution of association signals. Using a local ancestry-aware regression model, we replicate known hits for blood lipids, discover novel hits missed by standard GWAS and localize signals closer to putative causal variants.
Figures
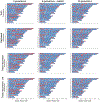
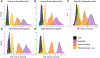


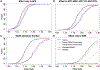

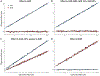
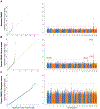

Comment in
-
Reply to: On powerful GWAS in admixed populations.Nat Genet. 2021 Dec;53(12):1634-1635. doi: 10.1038/s41588-021-00975-z. Epub 2021 Nov 25. Nat Genet. 2021. PMID: 34824479 No abstract available.
-
On powerful GWAS in admixed populations.Nat Genet. 2021 Dec;53(12):1631-1633. doi: 10.1038/s41588-021-00953-5. Epub 2021 Nov 25. Nat Genet. 2021. PMID: 34824480 Free PMC article. No abstract available.
References
-
- Parker K, Morin R, Horowitz Juliana Menasce & Rohal M Multiracial in America: Proud, Diverse and Growing in Numbers. (2015).
Online Methods References
-
- Van Rossum G, Drake FL Jr. Python reference manual. Centrum voor Wiskunde en Informatica; Amsterdam; 1995.
-
- Gnu P Bash (3.2.48) [Unix shell program; ]. 2007.
-
- Williams A. admix-simu: program to simulate admixture between multiple populations. 2016 doi: 10.5281/ZENODO.45517. . - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

