Protein arginine methyltransferase 5 (PRMT5) activates WNT/β-catenin signalling in breast cancer cells via epigenetic silencing of DKK1 and DKK3
- PMID: 33462997
- PMCID: PMC7875925
- DOI: 10.1111/jcmm.16260
Protein arginine methyltransferase 5 (PRMT5) activates WNT/β-catenin signalling in breast cancer cells via epigenetic silencing of DKK1 and DKK3
Abstract
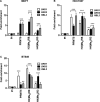
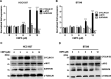
Protein arginine methyltransferase 5 (PRMT5) activity is dysregulated in many aggressive cancers and its enhanced levels are associated with increased tumour growth and survival. However, the role of PRMT5 in breast cancer remains underexplored. In this study, we show that PRMT5 is overexpressed in breast cancer cell lines, and that it promotes WNT/β-CATENIN proliferative signalling through epigenetic silencing of pathway antagonists, DKK1 and DKK3, leading to enhanced expression of c-MYC, CYCLIN D1 and SURVIVIN. Through chromatin immunoprecipitation (ChIP) studies, we found that PRMT5 binds to the promoter region of WNT antagonists, DKK1 and DKK3, and induces symmetric methylation of H3R8 and H4R3 histones. Our findings also show that PRMT5 inhibition using a specific small molecule inhibitor, compound 5 (CMP5), reduces PRMT5 recruitment as well as methylation of H3R8 and H4R3 histones in the promoter regions of DKK1 and DKK3, which consequently results in reduced expression CYCLIN D1 and SURVIVIN. Furthermore, CMP5 treatment either alone or in combination with 5-Azacytidine and Trichostatin A restored expression of DKK1 and DKK3 in TNBCs. PRMT5 inhibition also altered the growth characteristics of breast cancer cells and induced their death. Collectively, these results show that PRMT5 controls breast cancer cell growth through epigenetic silencing of WNT/β-CATENIN pathway antagonists, DKK1 and DKK3, resulting in up-regulation of WNT/β-CATENIN proliferative signalling.
Keywords: PRMT5; WNT/β-CATENIN proliferative signalling; breast cancer; epigenetic silencing; tumour suppressors.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest with the contents of this article.
Figures





References
-
- World Cancer Report 2014 . World Cancer Report. International Agency for Research on Cancer; 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

