Aqueous Processed Biopolymer Interfaces for Single-Cell Microarrays
- PMID: 33463257
- PMCID: PMC7997111
- DOI: 10.1021/acsbiomaterials.9b01871
Aqueous Processed Biopolymer Interfaces for Single-Cell Microarrays
Abstract
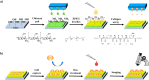
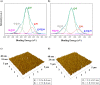
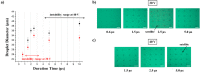
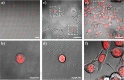
Single-cell microarrays are emerging tools to unravel intrinsic diversity within complex cell populations, opening up new approaches for the in-depth understanding of highly relevant diseases. However, most of the current methods for their fabrication are based on cumbersome patterning approaches, employing organic solvents and/or expensive materials. Here, we demonstrate an unprecedented green-chemistry strategy to produce single-cell capture biochips onto glass surfaces by all-aqueous inkjet printing. At first, a chitosan film is easily inkjet printed and immobilized onto hydroxyl-rich glass surfaces by electrostatic immobilization. In turn, poly(ethylene glycol) diglycidyl ether is grafted on the chitosan film to expose reactive epoxy groups and induce antifouling properties. Subsequently, microscale collagen spots are printed onto the above surface to define the attachment area for single adherent human cancer cells harvesting with high yield. The reported inkjet printing approach enables one to modulate the collagen area available for cell attachment in order to control the number of captured cells per spot, from single-cells up to double- and multiple-cell arrays. Proof-of-principle of the approach includes pharmacological treatment of single-cells by the model drug doxorubicin. The herein presented strategy for single-cell array fabrication can constitute a first step toward an innovative and environmentally friendly generation of aqueous-based inkjet-printed cellular devices.
Keywords: biointerface; biopolymer; inkjet printing; microarray; single-cell.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Inkjet-Printed Oxide Thin-Film Transistors Based on Nanopore-Free Aqueous-Processed Dielectric for Active-Matrix Quantum-Dot Light-Emitting Diode Displays.ACS Appl Mater Interfaces. 2019 Aug 7;11(31):28052-28059. doi: 10.1021/acsami.9b08258. Epub 2019 Jul 25. ACS Appl Mater Interfaces. 2019. PMID: 31304744
-
Protein microarray spots are modulated by patterning method, surface chemistry and processing conditions.Biosens Bioelectron. 2019 Apr 1;130:397-407. doi: 10.1016/j.bios.2018.09.027. Epub 2018 Sep 7. Biosens Bioelectron. 2019. PMID: 30253928
-
Cell micropatterning on an albumin-based substrate using an inkjet printing technique.J Biomed Mater Res A. 2009 Dec 15;91(4):1202-9. doi: 10.1002/jbm.a.32312. J Biomed Mater Res A. 2009. PMID: 19148930
-
Review of Recent Inkjet-Printed Capacitive Tactile Sensors.Sensors (Basel). 2017 Nov 10;17(11):2593. doi: 10.3390/s17112593. Sensors (Basel). 2017. PMID: 29125584 Free PMC article. Review.
-
Bio-microarray fabrication techniques--a review.Crit Rev Biotechnol. 2006 Oct-Dec;26(4):237-59. doi: 10.1080/07388550600978358. Crit Rev Biotechnol. 2006. PMID: 17095434 Review.
Cited by
-
Control of the Drying Patterns for Complex Colloidal Solutions and Their Applications.Nanomaterials (Basel). 2022 Jul 28;12(15):2600. doi: 10.3390/nano12152600. Nanomaterials (Basel). 2022. PMID: 35957030 Free PMC article. Review.
-
An electroactive hybrid biointerface for enhancing neuronal differentiation and axonal outgrowth on bio-subretinal chip.Mater Today Bio. 2022 Apr 5;14:100253. doi: 10.1016/j.mtbio.2022.100253. eCollection 2022 Mar. Mater Today Bio. 2022. PMID: 35464741 Free PMC article.
-
On the Interaction between 1D Materials and Living Cells.J Funct Biomater. 2020 Jun 10;11(2):40. doi: 10.3390/jfb11020040. J Funct Biomater. 2020. PMID: 32531950 Free PMC article. Review.
-
Understanding droplet jetting on varying substrate for biological applications.Int J Bioprint. 2023 May 23;9(5):758. doi: 10.18063/ijb.758. eCollection 2023. Int J Bioprint. 2023. PMID: 37457927 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
