Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e
- PMID: 33463525
- PMCID: PMC7861616
- DOI: 10.7554/eLife.64509
Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e
Erratum in
-
Correction: Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e.Elife. 2022 Sep 14;11:e83277. doi: 10.7554/eLife.83277. Elife. 2022. PMID: 36103315 Free PMC article.
Abstract
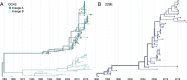
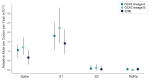
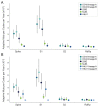
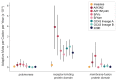
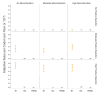
Seasonal coronaviruses (OC43, 229E, NL63, and HKU1) are endemic to the human population, regularly infecting and reinfecting humans while typically causing asymptomatic to mild respiratory infections. It is not known to what extent reinfection by these viruses is due to waning immune memory or antigenic drift of the viruses. Here we address the influence of antigenic drift on immune evasion of seasonal coronaviruses. We provide evidence that at least two of these viruses, OC43 and 229E, are undergoing adaptive evolution in regions of the viral spike protein that are exposed to human humoral immunity. This suggests that reinfection may be due, in part, to positively selected genetic changes in these viruses that enable them to escape recognition by the immune system. It is possible that, as with seasonal influenza, these adaptive changes in antigenic regions of the virus would necessitate continual reformulation of a vaccine made against them.
Keywords: 229e; adaptive evolution; antigenic evolution; evolutionary biology; oc43; seasonal coronavirus; virus.
© 2021, Kistler and Bedford.
Conflict of interest statement
KK, TB No competing interests declared
Figures






References
-
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, Matschiner M, Mendes FK, Müller NF, Ogilvie HA, du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu CH, Xie D, Zhang C, Stadler T, Drummond AJ. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLOS Computational Biology. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

