Phenotypic and molecular evolution across 10,000 generations in laboratory budding yeast populations
- PMID: 33464204
- PMCID: PMC7815316
- DOI: 10.7554/eLife.63910
Phenotypic and molecular evolution across 10,000 generations in laboratory budding yeast populations
Abstract
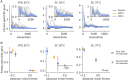
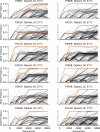
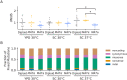
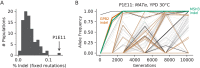
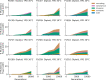
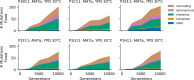
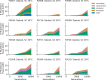
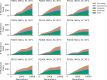
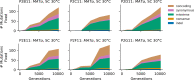
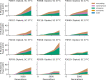
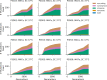
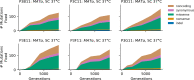
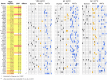
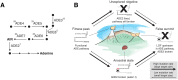
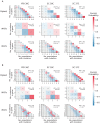
Laboratory experimental evolution provides a window into the details of the evolutionary process. To investigate the consequences of long-term adaptation, we evolved 205 Saccharomyces cerevisiae populations (124 haploid and 81 diploid) for ~10,000,000 generations in three environments. We measured the dynamics of fitness changes over time, finding repeatable patterns of declining adaptability. Sequencing revealed that this phenotypic adaptation is coupled with a steady accumulation of mutations, widespread genetic parallelism, and historical contingency. In contrast to long-term evolution in E. coli, we do not observe long-term coexistence or populations with highly elevated mutation rates. We find that evolution in diploid populations involves both fixation of heterozygous mutations and frequent loss-of-heterozygosity events. Together, these results help distinguish aspects of evolutionary dynamics that are likely to be general features of adaptation across many systems from those that are specific to individual organisms and environmental conditions.
Keywords: S. cerevisiae; dynamics of adaptation; evolutionary biology; experimental evolution; yeast.
© 2021, Johnson et al.
Conflict of interest statement
MJ, SG, JG, MD, CB, PH, TJ, EJ, KK, KL, JM, AM, AP, RP, AR, MM, AN, MD No competing interests declared, JP Julia C. Piper is affiliated with Aeronaut Brewing Co. The author has no financial interests to declare.
Figures




























Similar articles
-
Dynamics and variability in the pleiotropic effects of adaptation in laboratory budding yeast populations.Elife. 2021 Oct 1;10:e70918. doi: 10.7554/eLife.70918. Elife. 2021. PMID: 34596043 Free PMC article.
-
The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast.PLoS Genet. 2008 Dec;4(12):e1000303. doi: 10.1371/journal.pgen.1000303. Epub 2008 Dec 12. PLoS Genet. 2008. PMID: 19079573 Free PMC article.
-
The evolutionary plasticity of chromosome metabolism allows adaptation to constitutive DNA replication stress.Elife. 2020 Feb 11;9:e51963. doi: 10.7554/eLife.51963. Elife. 2020. PMID: 32043971 Free PMC article.
-
Loss of Heterozygosity and Its Importance in Evolution.J Mol Evol. 2023 Jun;91(3):369-377. doi: 10.1007/s00239-022-10088-8. Epub 2023 Feb 8. J Mol Evol. 2023. PMID: 36752826 Free PMC article. Review.
-
Revisiting Mortimer's Genome Renewal Hypothesis: heterozygosity, homothallism, and the potential for adaptation in yeast.Adv Exp Med Biol. 2014;781:37-48. doi: 10.1007/978-94-007-7347-9_3. Adv Exp Med Biol. 2014. PMID: 24277294 Free PMC article. Review.
Cited by
-
The genetic basis of differential autodiploidization in evolving yeast populations.G3 (Bethesda). 2021 Aug 7;11(8):jkab192. doi: 10.1093/g3journal/jkab192. G3 (Bethesda). 2021. PMID: 34849811 Free PMC article.
-
Identifying Targets of Selection in Laboratory Evolution Experiments.J Mol Evol. 2023 Jun;91(3):345-355. doi: 10.1007/s00239-023-10096-2. Epub 2023 Feb 21. J Mol Evol. 2023. PMID: 36810618 Free PMC article. Review.
-
Mutational robustness changes during long-term adaptation in laboratory budding yeast populations.Elife. 2022 Jul 26;11:e76491. doi: 10.7554/eLife.76491. Elife. 2022. PMID: 35880743 Free PMC article.
-
Competition dynamics in long-term propagations of Schizosaccharomyces pombe strain communities.Ecol Evol. 2021 Oct 9;11(21):15085-15097. doi: 10.1002/ece3.8191. eCollection 2021 Nov. Ecol Evol. 2021. PMID: 34765162 Free PMC article.
-
Recent insights into the evolution of mutation rates in yeast.Curr Opin Genet Dev. 2022 Oct;76:101953. doi: 10.1016/j.gde.2022.101953. Epub 2022 Jul 11. Curr Opin Genet Dev. 2022. PMID: 35834945 Free PMC article. Review.
References
-
- Belyeu JR, Brown J, Pedersen BS, Cormier MJ, Layer R, Brueffer C, Valle-Inclan JE. Samplot: A Platform for Structural Variant Visual Validation and Automated Filtering. 2fb0b75GitHub. 2020 https://github.com/ryanlayer/samplot - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

