LNCcation: lncRNA localization and function
- PMID: 33464299
- PMCID: PMC7816648
- DOI: 10.1083/jcb.202009045
LNCcation: lncRNA localization and function
Abstract
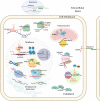
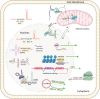
Subcellular localization of RNAs has gained attention in recent years as a prevalent phenomenon that influences numerous cellular processes. This is also evident for the large and relatively novel class of long noncoding RNAs (lncRNAs). Because lncRNAs are defined as RNA transcripts >200 nucleotides that do not encode protein, they are themselves the functional units, making their subcellular localization critical to their function. The discovery of tens of thousands of lncRNAs and the cumulative evidence involving them in almost every cellular activity render assessment of their subcellular localization essential to fully understanding their biology. In this review, we summarize current knowledge of lncRNA subcellular localization, factors controlling their localization, emerging themes, including the role of lncRNA isoforms and the involvement of lncRNAs in phase separation bodies, and the implications of lncRNA localization on their function and on cellular behavior. We also discuss gaps in the current knowledge as well as opportunities that these provide for novel avenues of investigation.
© 2021 Bridges et al.
Figures

References
-
- Amaral, P.P., Leonardi T., Han N., Viré E., Gascoigne D.K., Arias-Carrasco R., Büscher M., Pandolfini L., Zhang A., Pluchino S., et al. . 2018. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 19:32 10.1186/s13059-018-1405-5 - DOI - PMC - PubMed
-
- Anderson, D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., and Olson E.N.. 2015. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 160:595–606. 10.1016/j.cell.2015.01.009 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

