Hmx1 regulates urfh1 expression in the craniofacial region in zebrafish
- PMID: 33465110
- PMCID: PMC7815118
- DOI: 10.1371/journal.pone.0245239
Hmx1 regulates urfh1 expression in the craniofacial region in zebrafish
Abstract
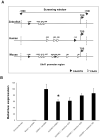
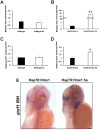
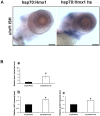
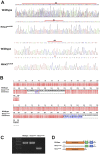
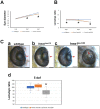
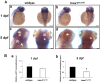
H6 family homeobox 1 (HMX1) regulates multiple aspects of craniofacial development as it is widely expressed in the eye, peripheral ganglia and branchial arches. Mutations in HMX1 are linked to an ocular defect termed Oculo-auricular syndrome of Schorderet-Munier-Franceschetti (MIM #612109). We identified UHRF1 as a target of HMX1 during development. UHRF1 and its partner proteins actively regulate chromatin modifications and cellular proliferation. Luciferase assays and in situ hybridization analyses showed that HMX1 exerts a transcriptional inhibitory effect on UHRF1 and a modification of its expression pattern. Overexpression of hmx1 in hsp70-hmx1 zebrafish increased uhrf1 expression in the cranial region, while mutations in the hmx1 dimerization domains reduced uhrf1 expression. Moreover, the expression level of uhrf1 and its partner dnmt1 was increased in the eye field in response to hmx1 overexpression. These results indicate that hmx1 regulates uhrf1 expression and, potentially through regulating the expression of factors involved in DNA methylation, contribute to the development of the craniofacial region of zebrafish.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

