Myogenin is required for assembly of the transcription machinery on muscle genes during skeletal muscle differentiation
- PMID: 33465133
- PMCID: PMC7815108
- DOI: 10.1371/journal.pone.0245618
Myogenin is required for assembly of the transcription machinery on muscle genes during skeletal muscle differentiation
Abstract
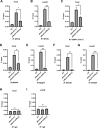
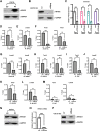
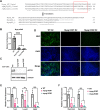
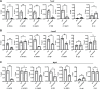
Skeletal muscle gene expression is governed by the myogenic regulatory family (MRF) which includes MyoD (MYOD1) and myogenin (MYOG). MYOD1 and MYOG are known to regulate an overlapping set of muscle genes, but MYOD1 cannot compensate for the absence of MYOG in vivo. In vitro, late muscle genes have been shown to be bound by both factors, but require MYOG for activation. The molecular basis for this requirement was unclear. We show here that MYOG is required for the recruitment of TBP and RNAPII to muscle gene promoters, indicating that MYOG is essential in assembling the transcription machinery. Genes regulated by MYOD1 and MYOG include genes required for muscle fusion, myomaker and myomerger, and we show that myomaker is fully dependent on activation by MYOG. We also sought to determine the role of MYOD1 in MYOG dependent gene activation and unexpectedly found that MYOG is required to maintain Myod1 expression. However, we also found that exogenous MYOD1 was unable to compensate for the loss of Myog and activate muscle gene expression. Thus, our results show that MYOD1 and MYOG act in a feed forward loop to maintain each other's expression and also show that it is MYOG, and not MYOD1, that is required to load TBP and activate gene expression on late muscle gene promoters bound by both factors.
Conflict of interest statement
The authors have declared that no competing interests exists.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

