Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2
- PMID: 33465165
- PMCID: PMC7845965
- DOI: 10.1371/journal.ppat.1009212
Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2
Abstract
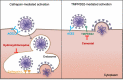
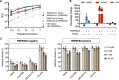
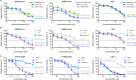
Hydroxychloroquine, used to treat malaria and some autoimmune disorders, potently inhibits viral infection of SARS coronavirus (SARS-CoV-1) and SARS-CoV-2 in cell-culture studies. However, human clinical trials of hydroxychloroquine failed to establish its usefulness as treatment for COVID-19. This compound is known to interfere with endosomal acidification necessary to the proteolytic activity of cathepsins. Following receptor binding and endocytosis, cathepsin L can cleave the SARS-CoV-1 and SARS-CoV-2 spike (S) proteins, thereby activating membrane fusion for cell entry. The plasma membrane-associated protease TMPRSS2 can similarly cleave these S proteins and activate viral entry at the cell surface. Here we show that the SARS-CoV-2 entry process is more dependent than that of SARS-CoV-1 on TMPRSS2 expression. This difference can be reversed when the furin-cleavage site of the SARS-CoV-2 S protein is ablated or when it is introduced into the SARS-CoV-1 S protein. We also show that hydroxychloroquine efficiently blocks viral entry mediated by cathepsin L, but not by TMPRSS2, and that a combination of hydroxychloroquine and a clinically-tested TMPRSS2 inhibitor prevents SARS-CoV-2 infection more potently than either drug alone. These studies identify functional differences between SARS-CoV-1 and -2 entry processes, and provide a mechanistic explanation for the limited in vivo utility of hydroxychloroquine as a treatment for COVID-19.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Dual Inhibition of Vacuolar-ATPase and TMPRSS2 Is Required for Complete Blockade of SARS-CoV-2 Entry into Cells.Antimicrob Agents Chemother. 2022 Jul 19;66(7):e0043922. doi: 10.1128/aac.00439-22. Epub 2022 Jun 15. Antimicrob Agents Chemother. 2022. PMID: 35703551 Free PMC article.
-
Synergistic Block of SARS-CoV-2 Infection by Combined Drug Inhibition of the Host Entry Factors PIKfyve Kinase and TMPRSS2 Protease.J Virol. 2021 Oct 13;95(21):e0097521. doi: 10.1128/JVI.00975-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406858 Free PMC article.
-
Reactive Centre Loop Mutagenesis of SerpinB3 to Target TMPRSS2 and Furin: Inhibition of SARS-CoV-2 Cell Entry and Replication.Int J Mol Sci. 2022 Oct 19;23(20):12522. doi: 10.3390/ijms232012522. Int J Mol Sci. 2022. PMID: 36293378 Free PMC article.
-
Targeting the viral-entry facilitators of SARS-CoV-2 as a therapeutic strategy in COVID-19.J Med Virol. 2021 Sep;93(9):5260-5276. doi: 10.1002/jmv.27019. Epub 2021 May 3. J Med Virol. 2021. PMID: 33851732 Free PMC article. Review.
-
Spiking dependence of SARS-CoV-2 pathogenicity on TMPRSS2.J Med Virol. 2021 Jul;93(7):4205-4218. doi: 10.1002/jmv.26911. Epub 2021 Mar 18. J Med Virol. 2021. PMID: 33638460 Free PMC article. Review.
Cited by
-
Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV-2-Priming Protease TMPRSS2.Pathog Immun. 2021 Apr 26;6(1):55-74. doi: 10.20411/pai.v6i1.408. eCollection 2021. Pathog Immun. 2021. PMID: 33969249 Free PMC article.
-
Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture.Commun Biol. 2022 Sep 14;5(1):958. doi: 10.1038/s42003-022-03841-8. Commun Biol. 2022. PMID: 36104427 Free PMC article.
-
Marine Alga Ulva fasciata-Derived Molecules for the Potential Treatment of SARS-CoV-2: An In Silico Approach.Mar Drugs. 2022 Sep 19;20(9):586. doi: 10.3390/md20090586. Mar Drugs. 2022. PMID: 36135775 Free PMC article.
-
Interferon-stimulated gene PVRL4 broadly suppresses viral entry by inhibiting viral-cellular membrane fusion.Cell Biosci. 2024 Feb 17;14(1):23. doi: 10.1186/s13578-024-01202-y. Cell Biosci. 2024. PMID: 38368366 Free PMC article.
-
Clinical drug therapies and biologicals currently used or in clinical trial to treat COVID-19.Biomed Pharmacother. 2021 Dec;144:112276. doi: 10.1016/j.biopha.2021.112276. Epub 2021 Oct 2. Biomed Pharmacother. 2021. PMID: 34624681 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

