Machine learning analysis using 77,044 genomic and transcriptomic profiles to accurately predict tumor type
- PMID: 33465745
- PMCID: PMC7815805
- DOI: 10.1016/j.tranon.2021.101016
Machine learning analysis using 77,044 genomic and transcriptomic profiles to accurately predict tumor type
Abstract
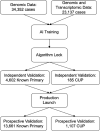
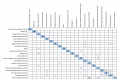
Cancer of Unknown Primary (CUP) occurs in 3-5% of patients when standard histological diagnostic tests are unable to determine the origin of metastatic cancer. Typically, a CUP diagnosis is treated empirically and has very poor outcomes, with median overall survival less than one year. Gene expression profiling alone has been used to identify the tissue of origin but struggles with low neoplastic percentage in metastatic sites which is where identification is often most needed. MI GPSai, a Genomic Prevalence Score, uses DNA sequencing and whole transcriptome data coupled with machine learning to aid in the diagnosis of cancer. The algorithm trained on genomic data from 34,352 cases and genomic and transcriptomic data from 23,137 cases and was validated on 19,555 cases. MI GPSai predicted the tumor type in the labeled data set with an accuracy of over 94% on 93% of cases while deliberating amongst 21 possible categories of cancer. When also considering the second highest prediction, the accuracy increases to 97%. Additionally, MI GPSai rendered a prediction for 71.7% of CUP cases. Pathologist evaluation of discrepancies between submitted diagnosis and MI GPSai predictions resulted in change of diagnosis in 41.3% of the time. MI GPSai provides clinically meaningful information in a large proportion of CUP cases and inclusion of MI GPSai in clinical routine could improve diagnostic fidelity. Moreover, all genomic markers essential for therapy selection are assessed in this assay, maximizing the clinical utility for patients within a single test.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Massard C., Loriot Y., Fizazi K. Carcinomas of an unknown primary origin–diagnosis and treatment. Nat. Rev. Clin. Oncol. 2011;8(12):701–710. - PubMed
-
- Varadhachary G.R., Raber M.N. Cancer of unknown primary site. N. Engl. J. Med. 2014;371(8):757–765. - PubMed
-
- DeYoung B.R., Wick M.R. Immunohistologic evaluation of metastatic carcinomas of unknown origin: an algorithmic approach. Semin. Diagn. Pathol. 2000;17(3):184–193. - PubMed
-
- Anderson G.G., Weiss L.M. Determining tissue of origin for metastatic cancers: meta-analysis and literature review of immunohistochemistry performance. Appl. Immunohistochem. Mol. Morphol. 2010;18(1):3–8. - PubMed
-
- Park S.Y., Kim B.H., Kim J.H., Lee S., Kang G.H. Panels of immunohistochemical markers help determine primary sites of metastatic adenocarcinoma. Arch. Pathol. Lab. Med. 2007;131(10):1561–1567. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

