Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models
- PMID: 33466285
- PMCID: PMC7824762
- DOI: 10.3390/cells10010071
Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models
Abstract
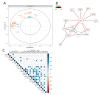
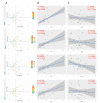
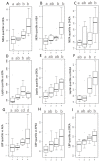
The global obesity epidemic continues to rise worldwide. In this context, unraveling new interconnections between biological systems involved in obesity etiology is highly relevant. Dysregulation of the endocannabinoidome (eCBome) is associated with metabolic complications in obesity. This study aims at deciphering new associations between circulating endogenous bioactive lipids belonging to the eCBome and metabolic parameters in a population of overweight or obese individuals with metabolic syndrome. To this aim, we combined different multivariate exploratory analysis methods: canonical correlation analysis and principal component analysis, revealed associations between eCBome subsets, and metabolic parameters such as leptin, lipopolysaccharide-binding protein, and non-esterified fatty acids (NEFA). Subsequent construction of predictive regression models according to the linear combination of selected endocannabinoids demonstrates good prediction performance for NEFA. Descriptive approaches reveal the importance of specific circulating endocannabinoids and key related congeners to explain variance in the metabolic parameters in our cohort. Analysis of quartiles confirmed that these bioactive lipids were significantly higher in individuals characterized by important levels for aforementioned metabolic variables. In conclusion, by proposing a methodology for the exploration of large-scale data, our study offers additional evidence of the existence of an interplay between eCBome related-entities and metabolic parameters known to be altered in obesity.
Trial registration: ClinicalTrials.gov NCT02637115.
Keywords: LBP; NEFA; endocannabinoidome; endocannabinoids; human; leptin; metabolic syndrome; multivariate analysis; obesity.
Conflict of interest statement
P.D.C. is co-founder of A-Mansia Biotech SA. The other authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

