Kinetic Modeling and Meta-Analysis of the Bacillus subtilis SigB Regulon during Spore Germination and Outgrowth
- PMID: 33466511
- PMCID: PMC7824861
- DOI: 10.3390/microorganisms9010112
Kinetic Modeling and Meta-Analysis of the Bacillus subtilis SigB Regulon during Spore Germination and Outgrowth
Abstract
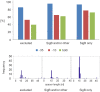
The exponential increase in the number of conducted studies combined with the development of sequencing methods have led to an enormous accumulation of partially processed experimental data in the past two decades. Here, we present an approach using literature-mined data complemented with gene expression kinetic modeling and promoter sequence analysis. This approach allowed us to identify the regulon of Bacillus subtilis sigma factor SigB of RNA polymerase (RNAP) specifically expressed during germination and outgrowth. SigB is critical for the cell's response to general stress but is also expressed during spore germination and outgrowth, and this specific regulon is not known. This approach allowed us to (i) define a subset of the known SigB regulon controlled by SigB specifically during spore germination and outgrowth, (ii) identify the influence of the promoter sequence binding motif organization on the expression of the SigB-regulated genes, and (iii) suggest additional sigma factors co-controlling other SigB-dependent genes. Experiments then validated promoter sequence characteristics necessary for direct RNAP-SigB binding. In summary, this work documents the potential of computational approaches to unravel new information even for a well-studied system; moreover, the study specifically identifies the subset of the SigB regulon, which is activated during germination and outgrowth.
Keywords: Bacillus subtilis; SigB; computational modeling; gene regulatory networks; promoter sequence analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
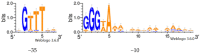

Similar articles
-
Quantitative Aspect of Bacillus subtilis σB Regulatory Network-A Computational Simulation.Biology (Basel). 2022 Nov 29;11(12):1729. doi: 10.3390/biology11121729. Biology (Basel). 2022. PMID: 36552239 Free PMC article.
-
Prediction and validation of novel SigB regulon members in Bacillus subtilis and regulon structure comparison to Bacillales members.BMC Microbiol. 2023 Jan 18;23(1):17. doi: 10.1186/s12866-022-02700-0. BMC Microbiol. 2023. PMID: 36653740 Free PMC article.
-
The Stress-Responsive Alternative Sigma Factor SigB of Bacillus subtilis and Its Relatives: An Old Friend With New Functions.Front Microbiol. 2020 Sep 15;11:1761. doi: 10.3389/fmicb.2020.01761. eCollection 2020. Front Microbiol. 2020. PMID: 33042030 Free PMC article. Review.
-
Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification.Microbiology (Reading). 2012 Mar;158(Pt 3):696-707. doi: 10.1099/mic.0.055434-0. Epub 2011 Dec 15. Microbiology (Reading). 2012. PMID: 22174379
-
Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon.Mol Microbiol. 1998 Sep;29(5):1129-36. doi: 10.1046/j.1365-2958.1998.00977.x. Mol Microbiol. 1998. PMID: 9767581 Review.
Cited by
-
MoaB2, a newly identified transcription factor, binds to σA in Mycobacterium smegmatis.J Bacteriol. 2024 Dec 19;206(12):e0006624. doi: 10.1128/jb.00066-24. Epub 2024 Nov 5. J Bacteriol. 2024. PMID: 39499088 Free PMC article.
-
σE of Streptomyces coelicolor can function both as a direct activator or repressor of transcription.Commun Biol. 2024 Jan 6;7(1):46. doi: 10.1038/s42003-023-05716-y. Commun Biol. 2024. PMID: 38184746 Free PMC article.
-
Quantitative Aspect of Bacillus subtilis σB Regulatory Network-A Computational Simulation.Biology (Basel). 2022 Nov 29;11(12):1729. doi: 10.3390/biology11121729. Biology (Basel). 2022. PMID: 36552239 Free PMC article.
-
Special Issue "Bacillus subtilis as a Model Organism to Study Basic Cell Processes".Microorganisms. 2021 Nov 28;9(12):2459. doi: 10.3390/microorganisms9122459. Microorganisms. 2021. PMID: 34946061 Free PMC article.
-
Understanding the Effect of Different Glucose Concentrations in the Oligotrophic Bacterium Bacillus subtilis BS-G1 through Transcriptomics Analysis.Microorganisms. 2023 Sep 26;11(10):2401. doi: 10.3390/microorganisms11102401. Microorganisms. 2023. PMID: 37894061 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

