AtPiezo Plays an Important Role in Root Cap Mechanotransduction
- PMID: 33466520
- PMCID: PMC7796506
- DOI: 10.3390/ijms22010467
AtPiezo Plays an Important Role in Root Cap Mechanotransduction
Abstract
Plants encounter a variety of mechanical stimuli during their growth and development. It is currently believed that mechanosensitive ion channels play an essential role in the initial perception of mechanical force in plants. Over the past decade, the study of Piezo, a mechanosensitive ion channel in animals, has made significant progress. It has been proved that the perception of mechanical force in various physiological processes of animals is indispensable. However, little is still known about the function of its homologs in plants. In this study, by investigating the function of the AtPiezo gene in the model plant Arabidopsis thaliana, we found that AtPiezo plays a role in the perception of mechanical force in plant root cap and the flow of Ca2+ is involved in this process. These findings allow us to understand the function of AtPiezo from the perspective of plants and provide new insights into the mechanism of plant root cap in response to mechanical stimuli.
Keywords: Arabidopsis; AtPiezo; Ca2+; mechanical stimuli; mechanosensitive ion channel; root cap.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

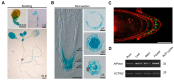

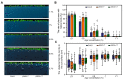
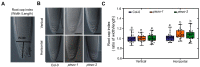
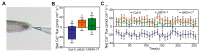
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

