Synergism of Proneurogenic miRNAs Provides a More Effective Strategy to Target Glioma Stem Cells
- PMID: 33466745
- PMCID: PMC7831004
- DOI: 10.3390/cancers13020289
Synergism of Proneurogenic miRNAs Provides a More Effective Strategy to Target Glioma Stem Cells
Abstract
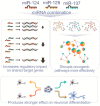
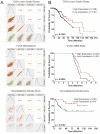
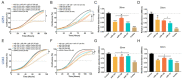
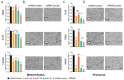
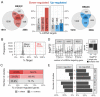
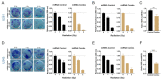
Tumor suppressor microRNAs (miRNAs) have been explored as agents to target cancer stem cells. Most strategies use a single miRNA mimic and present many disadvantages, such as the amount of reagent required and the diluted effect on target genes. miRNAs work in a cooperative fashion to regulate distinct biological processes and pathways. Therefore, we propose that miRNA combinations could provide more efficient ways to target cancer stem cells. We have previously shown that miR-124, miR-128, and miR-137 function synergistically to regulate neurogenesis. We used a combination of these three miRNAs to treat glioma stem cells and showed that this treatment was much more effective than single miRNAs in disrupting cell proliferation and survival and promoting differentiation and response to radiation. Transcriptomic analyses indicated that transcription regulation, angiogenesis, metabolism, and neuronal differentiation are among the main biological processes affected by transfection of this miRNA combination. In conclusion, we demonstrated the value of using combinations of neurogenic miRNAs to disrupt cancer phenotypes and glioma stem cell growth. The synergistic effect of these three miRNA amplified the repression of oncogenic factors and the effect on cancer relevant pathways. Future therapeutic approaches would benefit from utilizing miRNA combinations, especially when targeting cancer-initiating cell populations.
Keywords: glioblastoma; miR-124; miR-128; miR-137; miRNA; neuroblastoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
miR-124, -128, and -137 Orchestrate Neural Differentiation by Acting on Overlapping Gene Sets Containing a Highly Connected Transcription Factor Network.Stem Cells. 2016 Jan;34(1):220-32. doi: 10.1002/stem.2204. Epub 2015 Sep 15. Stem Cells. 2016. PMID: 26369286
-
Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review.
-
MiRNAs Mediate GDNF-Induced Proliferation and Migration of Glioma Cells.Cell Physiol Biochem. 2017;44(5):1923-1938. doi: 10.1159/000485883. Epub 2017 Dec 8. Cell Physiol Biochem. 2017. PMID: 29224008
-
Peritoneal fluid modifies the microRNA expression profile in endometrial and endometriotic cells from women with endometriosis.Hum Reprod. 2015 Oct;30(10):2292-302. doi: 10.1093/humrep/dev204. Epub 2015 Aug 25. Hum Reprod. 2015. PMID: 26307093
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
LncRNA FOXD3-AS1 Promotes Tumorigenesis of Glioma via Targeting miR-128-3p/SZRD1 Axis.Cancer Manag Res. 2021 Dec 7;13:9037-9048. doi: 10.2147/CMAR.S324920. eCollection 2021. Cancer Manag Res. 2021. Retraction in: Cancer Manag Res. 2022 Jan 20;14:257-258. doi: 10.2147/CMAR.S358886. PMID: 34916848 Free PMC article. Retracted.
-
Regulatory Functions of microRNAs in Cancer Stem Cells: Mechanism, Facts, and Perspectives.Cells. 2025 Jul 14;14(14):1073. doi: 10.3390/cells14141073. Cells. 2025. PMID: 40710326 Free PMC article. Review.
-
Exploring miRNA therapies and gut microbiome-enhanced CAR-T cells: advancing frontiers in glioblastoma stem cell targeting.Naunyn Schmiedebergs Arch Pharmacol. 2025 Mar;398(3):2169-2207. doi: 10.1007/s00210-024-03479-9. Epub 2024 Oct 9. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 39382681 Review.
-
ELF4 is a critical component of a miRNA-transcription factor network and is a bridge regulator of glioblastoma receptor signaling and lipid dynamics.Neuro Oncol. 2023 Mar 14;25(3):459-470. doi: 10.1093/neuonc/noac179. Neuro Oncol. 2023. PMID: 35862252 Free PMC article.
-
A miR-137-XIAP axis contributes to the sensitivity of TRAIL-induced cell death in glioblastoma.Front Oncol. 2022 Jul 28;12:870034. doi: 10.3389/fonc.2022.870034. eCollection 2022. Front Oncol. 2022. PMID: 35965517 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

