A Genotype-Phenotype Correlation Study of Exon Skip-Equivalent In-Frame Deletions and Exon Skip-Amenable Out-of-Frame Deletions across the DMD Gene to Simulate the Effects of Exon-Skipping Therapies: A Meta-Analysis
- PMID: 33466756
- PMCID: PMC7830903
- DOI: 10.3390/jpm11010046
A Genotype-Phenotype Correlation Study of Exon Skip-Equivalent In-Frame Deletions and Exon Skip-Amenable Out-of-Frame Deletions across the DMD Gene to Simulate the Effects of Exon-Skipping Therapies: A Meta-Analysis
Abstract
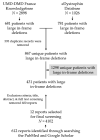
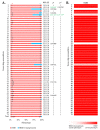
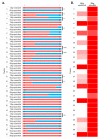
Dystrophinopathies are caused by mutations in the DMD gene. Out-of-frame deletions represent most mutational events in severe Duchenne muscular dystrophy (DMD), while in-frame deletions typically lead to milder Becker muscular dystrophy (BMD). Antisense oligonucleotide-mediated exon skipping converts an out-of-frame transcript to an in-frame one, inducing a truncated but partially functional dystrophin protein. The reading frame rule, however, has many exceptions. We thus sought to simulate clinical outcomes of exon-skipping therapies for DMD exons from clinical data of exon skip-equivalent in-frame deletions, in which the expressed quasi-dystrophins are comparable to those resulting from exon-skipping therapies. We identified a total of 1298 unique patients with exon skip-equivalent mutations in patient registries and the existing literature. We classified them into skip-equivalent deletions of each exon and statistically compared the ratio of DMD/BMD and asymptomatic individuals across the DMD gene. Our analysis identified that five exons are associated with significantly milder phenotypes than all other exons when corresponding exon skip-equivalent in-frame deletion mutations occur. Most exon skip-equivalent in-frame deletions were associated with a significantly milder phenotype compared to corresponding exon skip-amenable out-of-frame mutations. This study indicates the importance of genotype-phenotype correlation studies in the rational design of exon-skipping therapies.
Keywords: becker muscular dystrophy (BMD); duchenne muscular dystrophy (DMD); dystrophin; dystrophinopathy; exon skipping; reading frame rule; skip-equivalent deletions.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Koenig M., Beggs A.H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Müller C.R., Lindlöf M., Kaariainen H., et al. The molecular basis for duchenne versus becker muscular dystrophy: Correlation of severity with type of deletion. Am. J. Hum. Genet. 1989;45:498–506. doi: 10.1016/1. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

