Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing
- PMID: 33467183
- PMCID: PMC7830261
- DOI: 10.3390/genes12010106
Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing
Abstract
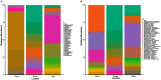
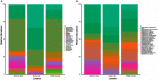
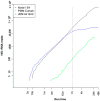
For the past two decades, microbial monitoring of the International Space Station (ISS) has relied on culture-dependent methods that require return to Earth for analysis. This has a number of limitations, with the most significant being bias towards the detection of culturable organisms and the inherent delay between sample collection and ground-based analysis. In recent years, portable and easy-to-use molecular-based tools, such as Oxford Nanopore Technologies' MinION™ sequencer and miniPCR bio's miniPCR™ thermal cycler, have been validated onboard the ISS. Here, we report on the development, validation, and implementation of a swab-to-sequencer method that provides a culture-independent solution to real-time microbial profiling onboard the ISS. Method development focused on analysis of swabs collected in a low-biomass environment with limited facility resources and stringent controls on allowed processes and reagents. ISS-optimized procedures included enzymatic DNA extraction from a swab tip, bead-based purifications, altered buffers, and the use of miniPCR and the MinION. Validation was conducted through extensive ground-based assessments comparing current standard culture-dependent and newly developed culture-independent methods. Similar microbial distributions were observed between the two methods; however, as expected, the culture-independent data revealed microbial profiles with greater diversity. Protocol optimization and verification was established during NASA Extreme Environment Mission Operations (NEEMO) analog missions 21 and 22, respectively. Unique microbial profiles obtained from analog testing validated the swab-to-sequencer method in an extreme environment. Finally, four independent swab-to-sequencer experiments were conducted onboard the ISS by two crewmembers. Microorganisms identified from ISS swabs were consistent with historical culture-based data, and primarily consisted of commonly observed human-associated microbes. This simplified method has been streamlined for high ease-of-use for a non-trained crew to complete in an extreme environment, thereby enabling environmental and human health diagnostics in real-time as future missions take us beyond low-Earth orbit.
Keywords: bacterial identification; field-deployable methods; in-situ analysis; nanopore sequencing; spaceflight.
Conflict of interest statement
S.J., D.J.T., and D.S. are employed by Oxford Nanopore Technologies, which manufactures and markets the MinION sequencing technology. These two authors supported method development. Other authors declare no conflict of interest.
Figures




References
-
- Pierson D.L. Microbial contamination of spacecraft. Gravitat. Space Biol. Bull. 2001;14:1–6. - PubMed
-
- Ferguson J.K., Taylor G.R., Mieszkuc B.J. Microbiological Investigations. In: Johnston R.S., Dietlein L.F., Berry C.A., editors. Biomedical Results of Apollo. National Aeronautics and Space Administration; Washington, DC, USA: 1975.
-
- Taylor G.R., Graves R.C., Brockett R.M., Ferguson J.K., Mieszuc B.J. Skylab Environmental and Crew Microbiology Studies. In: Johnston R.S., Dietlein L.F., editors. Biomedical Results from Skylab. National Aeronautics and Space Administration; Washington, DC, USA: 1977.
-
- Pierson D.B., Ott C.M., Bruce R., Castro A.V., Mehta S.K. American Institute of Aeronautics and Astronautics Meeting Papers, Proceedings of the 41st International Conference on Environmental Systems, International Conference on Environmental Systems (ICES), Portland, OR, USA, 17 July 2011. American Institute of Aeronautics and Astronautics; Reston, VA, USA: 2012. Microbiological Lessons Learned From the Space Shuttle; p. 5266.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

