MicroRNAs for Virus Pathogenicity and Host Responses, Identified in SARS-CoV-2 Genomes, May Play Roles in Viral-Host Co-Evolution in Putative Zoonotic Host Species
- PMID: 33467206
- PMCID: PMC7830670
- DOI: 10.3390/v13010117
MicroRNAs for Virus Pathogenicity and Host Responses, Identified in SARS-CoV-2 Genomes, May Play Roles in Viral-Host Co-Evolution in Putative Zoonotic Host Species
Abstract
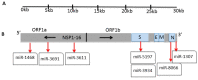
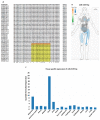
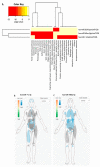
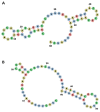
Our recent study identified seven key microRNAs (miR-8066, 5197, 3611, 3934-3p, 1307-3p, 3691-3p, 1468-5p) similar between SARS-CoV-2 and the human genome, pointing at miR-related mechanisms in viral entry and the regulatory effects on host immunity. To identify the putative roles of these miRs in zoonosis, we assessed their conservation, compared with humans, in some key wild and domestic animal carriers of zoonotic viruses, including bat, pangolin, pig, cow, rat, and chicken. Out of the seven miRs under study, miR-3611 was the most strongly conserved across all species; miR-5197 was the most conserved in pangolin, pig, cow, bat, and rat; miR-1307 was most strongly conserved in pangolin, pig, cow, bat, and human; miR-3691-3p in pangolin, cow, and human; miR-3934-3p in pig and cow, followed by pangolin and bat; miR-1468 was most conserved in pangolin, pig, and bat; while miR-8066 was most conserved in pangolin and pig. In humans, miR-3611 and miR-1307 were most conserved, while miR-8066, miR-5197, miR-3334-3p and miR-1468 were least conserved, compared with pangolin, pig, cow, and bat. Furthermore, we identified that changes in the miR-5197 nucleotides between pangolin and human can generate three new miRs, with differing tissue distribution in the brain, lung, intestines, lymph nodes, and muscle, and with different downstream regulatory effects on KEGG pathways. This may be of considerable importance as miR-5197 is localized in the spike protein transcript area of the SARS-CoV-2 genome. Our findings may indicate roles for these miRs in viral-host co-evolution in zoonotic hosts, particularly highlighting pangolin, bat, cow, and pig as putative zoonotic carriers, while highlighting the miRs' roles in KEGG pathways linked to viral pathogenicity and host responses in humans. This in silico study paves the way for investigations into the roles of miRs in zoonotic disease.
Keywords: COVID-19; SARS-CoV-2; co-evolution; microRNA; viral pathogenesis; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Driggin E., Madhavan M.V., Bikdeli B., Chuich T., Laracy J., Biondi-Zoccai G., Brown T.S., Der Nigoghossian C., Zidar D.A., Haythe J., et al. Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems During the Coronavirus Disease 2019 (COVID-19) Pandemic. J. Am. Coll. Cardiol. 2020;75:2352–2371. doi: 10.1016/j.jacc.2020.03.031. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

