The Proteome and Citrullinome of Hippoglossus hippoglossus Extracellular Vesicles-Novel Insights into Roles of the Serum Secretome in Immune, Gene Regulatory and Metabolic Pathways
- PMID: 33467210
- PMCID: PMC7830382
- DOI: 10.3390/ijms22020875
The Proteome and Citrullinome of Hippoglossus hippoglossus Extracellular Vesicles-Novel Insights into Roles of the Serum Secretome in Immune, Gene Regulatory and Metabolic Pathways
Abstract
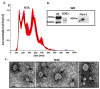
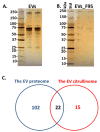
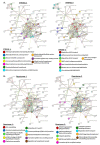
Extracellular vesicles (EVs) are lipid bilayer vesicles which are released from cells and play multifaceted roles in cellular communication in health and disease. EVs can be isolated from various body fluids, including serum and plasma, and are usable biomarkers as they can inform health status. Studies on EVs are an emerging research field in teleost fish, with accumulating evidence for important functions in immunity and homeostasis, but remain to be characterised in most fish species, including halibut. Protein deimination is a post-translational modification caused by a conserved family of enzymes, named peptidylarginine deiminases (PADs), and results in changes in protein folding and function via conversion of arginine to citrulline in target proteins. Protein deimination has been recently described in halibut ontogeny and halibut serum. Neither EV profiles, nor total protein or deiminated protein EV cargos have yet been assessed in halibut and are reported in the current study. Halibut serum EVs showed a poly-dispersed population in the size range of 50-600 nm, with modal size of EVs falling at 138 nm, and morphology was further confirmed by transmission electron microscopy. The assessment of EV total protein cargo revealed 124 protein hits and 37 deiminated protein hits, whereof 15 hits were particularly identified in deiminated form only. Protein interaction network analysis showed that deimination hits are involved in a range of gene regulatory, immune, metabolic and developmental processes. The same was found for total EV protein cargo, although a far wider range of pathways was found than for deimination hits only. The expression of complement component C3 and C4, as well as pentraxin-like protein, which were identified by proteomic analysis, was further verified in EVs by western blotting. This showed that C3 is exported in EVs at higher levels than C4 and deiminated C3 was furthermore confirmed to be at high levels in the deimination-enriched EV fractions, while, in comparison, C4 showed very low detection in deimination-enriched EV fractions. Pentraxin was exported in EVs, but not detected in the deimination-enriched fractions. Our findings provide novel insights into EV-mediated communication in halibut serum, via transport of protein cargo, including post-translationally deiminated proteins.
Keywords: citrullinome; complement; deimination/citrullination; extracellular vesicles; gene regulation; immunity; metabolism; pentraxin; peptidylarginine deiminase; proteome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






References
-
- Russel F.S. The Eggs and Planktonic Stages of British Marine Fishes. Academic Press; London, UK: 1976.
-
- Mangor-Jensen A., Harboe T., Shields R.J., Gara B., Naas K.E. Atlantic halibut, Hippoglossus hippoglossus L.; larvae cultivation literature, including a bibliography. Aquac. Res. 1998;29:857–886. doi: 10.1046/j.1365-2109.1998.29120857.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

