Chlamydia Uses K+ Electrical Signalling to Orchestrate Host Sensing, Inter-Bacterial Communication and Differentiation
- PMID: 33467438
- PMCID: PMC7830353
- DOI: 10.3390/microorganisms9010173
Chlamydia Uses K+ Electrical Signalling to Orchestrate Host Sensing, Inter-Bacterial Communication and Differentiation
Abstract
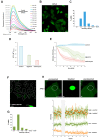
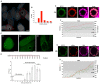
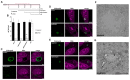
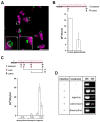
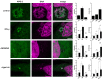
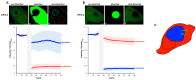
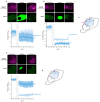
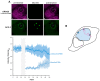
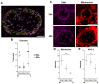
Prokaryotic communities coordinate quorum behaviour in response to external stimuli to control fundamental processes including inter-bacterial communication. The obligate intracellular bacterial pathogen Chlamydia adopts two developmental forms, invasive elementary bodies (EBs) and replicative reticulate bodies (RBs), which reside within a specialised membrane-bound compartment within the host cell termed an inclusion. The mechanisms by which this bacterial community orchestrates different stages of development from within the inclusion in coordination with the host remain elusive. Both prokaryotic and eukaryotic kingdoms exploit ion-based electrical signalling for fast intercellular communication. Here we demonstrate that RBs specifically accumulate potassium (K+) ions, generating a gradient. Disruption of this gradient using ionophores or an ion-channel inhibitor stalls the Chlamydia lifecycle, inducing persistence. Using photobleaching approaches, we establish that the RB is the master regulator of this [K+] differential and observe a fast K+ exchange between RBs revealing a role for this ion in inter-bacterial communication. Finally, we demonstrate spatio-temporal regulation of bacterial membrane potential during RB to EB differentiation within the inclusion. Together, our data reveal that Chlamydia harnesses K+ to orchestrate host sensing, inter-bacteria communication and pathogen differentiation.
Keywords: Chlamydia; cell-to-cell communications and community; host-pathogen interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Eley A. Chlamydia trachomatis is bad for your sperm! Microbiol. Today. 2003;30:61–62.
-
- Skowasch D., Yeghiazaryan K., Schrempf S., Golubnitschaja O., Welsch U., Preusse C.J., A Likungu J., Welz A., Lüderitz B., Bauriedel G. Persistence of Chlamydia pneumoniae in degenerative aortic valve stenosis indicated by heat shock protein 60 homologues. J. Heart Valve Dis. 2003;12:68–75. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

