Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis in Corylus heterophylla Fisch. × Corylus avellana L
- PMID: 33467497
- PMCID: PMC7830083
- DOI: 10.3390/plants10010159
Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis in Corylus heterophylla Fisch. × Corylus avellana L
Abstract
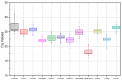
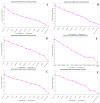
(1) Background: the species of Corylus have sporophytic type of self-incompatibility. Several genes related to recognition reaction between pollen and stigma have been identified in hazelnuts. To better understand the self-incompatibility (SI) response, we screened the suitable reference genes by using quantitative real-time reverse transcription PCR (qRT-PCR) analysis in hazelnut for the first time. (2) Methods: the major cultivar "Dawei" was used as material. A total of 12 candidate genes were identified and their expression profiles were compared among different tissues and in response to various treatments (different times after self- and cross-pollination) by RT-qPCR. The expression stability of these 12 candidate reference genes was evaluated using geNorm, NormFinder, BestKeeper, Delta Ct, and RefFinder programs. (3) Results: the comprehensive ranking of RefFinder indicated that ChaActin, VvActin, ChaUBQ14, and ChaEF1-α were the most suitable reference genes. According to the stability analysis of 12 candidate reference genes for each sample group based on four software packages, ChaActin and ChaEF1-α were most stable in different times after self-pollination and 4 h after self- and cross-pollination, respectively. To further validate the suitability of the reference genes identified in this study, CavPrx, which the expression profiles in Corylus have been reported, was quantified by using ChaActin and ChaEF1-α as reference genes. (4) Conclusions: our study of reference genes selection in hazelnut shows that the two reference genes, ChaActin and ChaEF1-α, are suitable for the evaluation of gene expression, and can be used for the analysis of pollen-pistil interaction in Corylus. The results supply a reliable foundation for accurate gene quantifications in Corylus species, which will facilitate the studies related to the reproductive biology in Corylus.
Keywords: Ping’ou hybrid hazelnut (C. heterophylla Fisch. × C. avellana L.); real-time quantitative PCR; reference gene; self-incompatibility; stability of gene expression.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Molecular cloning and yeast two-hybrid provide new evidence for unique sporophytic self-incompatibility system of Corylus.Plant Biol (Stuttg). 2022 Jan;24(1):104-116. doi: 10.1111/plb.13347. Epub 2021 Nov 1. Plant Biol (Stuttg). 2022. PMID: 34724309
-
Stigmatic Transcriptome Analysis of Self-Incompatible and Compatible Pollination in Corylus heterophylla Fisch. × Corylus avellana L.Front Plant Sci. 2022 Mar 1;13:800768. doi: 10.3389/fpls.2022.800768. eCollection 2022. Front Plant Sci. 2022. PMID: 35300011 Free PMC article.
-
Molecular cloning and expression analysis of hybrid hazelnut (Corylus heterophylla × Corylus avellana) ChaSRK1/2 genes and their homologs from other cultivars and species.Gene. 2020 Sep 25;756:144917. doi: 10.1016/j.gene.2020.144917. Epub 2020 Jun 23. Gene. 2020. PMID: 32590104
-
Final report on the safety assessment of Corylus Avellana (Hazel) Seed Oil, Corylus Americana (Hazel) Seed Oil, Corylus Avellana (Hazel) Seed Extract, Corylus Americana (Hazel) Seed Extract, Corylus Avellana (Hazel) Leaf Extract, Corylus Americana (Hazel) Leaf Extract, and Corylus Rostrata (Hazel) Leaf Extract.Int J Toxicol. 2001;20 Suppl 1:15-20. doi: 10.1080/109158101750300928. Int J Toxicol. 2001. PMID: 11358108 Review.
-
Multipurpose plant species and circular economy: Corylus avellana L. as a study case.Front Biosci (Landmark Ed). 2022 Jan 11;27(1):11. doi: 10.31083/j.fbl2701011. Front Biosci (Landmark Ed). 2022. PMID: 35090316 Review.
Cited by
-
Selection and validation of appropriate reference genes for RT-qPCR analysis of Nitraria sibirica under various abiotic stresses.BMC Plant Biol. 2022 Dec 17;22(1):592. doi: 10.1186/s12870-022-03988-w. BMC Plant Biol. 2022. PMID: 36526980 Free PMC article.
-
Identification and validation of reference genes for qPCR of Pseudomonas aeruginosa L10 under varying n-hexadecane concentrations.BMC Microbiol. 2025 Jul 2;25(1):390. doi: 10.1186/s12866-025-04112-2. BMC Microbiol. 2025. PMID: 40604373 Free PMC article.
-
Selection of Suitable Reference Genes for Gene Expression Normalization Studies in Dendrobium huoshanense.Genes (Basel). 2022 Aug 19;13(8):1486. doi: 10.3390/genes13081486. Genes (Basel). 2022. PMID: 36011396 Free PMC article.
-
Metabolomics and WGCNA Analyses Reveal the Underlying Mechanisms of Resistance to Botrytis cinerea in Hazelnut.Genes (Basel). 2024 Dec 24;16(1):2. doi: 10.3390/genes16010002. Genes (Basel). 2024. PMID: 39858549 Free PMC article.
-
Selection and Verification of Standardized Reference Genes of Angelica dahurica under Various Abiotic Stresses by Real-Time Quantitative PCR.Genes (Basel). 2024 Jan 7;15(1):79. doi: 10.3390/genes15010079. Genes (Basel). 2024. PMID: 38254968 Free PMC article.
References
-
- Boccacci P., Beltramo C., Sandoval Prando M.A., Lembo A., Sartor C., Mehlenbacher S.A., Torello Marinoni D. In silico mining, characterization and cross-species transferability of EST-SSR markers for European hazelnut (Corylus avellana L.) Mol. Breed. 2015;35:21. doi: 10.1007/s11032-015-0195-7. - DOI
-
- Mehlenbacher S.A. Advances in genetic improvement of hazelnut. Acta Hortic. 2018;1226:12. doi: 10.17660/ActaHortic.2018.1226.1. - DOI
-
- Henry R.J. Forest Trees. Springer; Berlin, Germany: 2011. Wild Crop Relatives: Genomic and Breeding Resources.
-
- Mehlenbacher S.A. Genetic resources for hazelnut: State of the art and future perspectives. Acta Hortic. 2009;845:33–38. doi: 10.17660/ActaHortic.2009.845.1. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

