Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis in Corylus heterophylla Fisch. × Corylus avellana L
- PMID: 33467497
- PMCID: PMC7830083
- DOI: 10.3390/plants10010159
Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis in Corylus heterophylla Fisch. × Corylus avellana L
Abstract
(1) Background: the species of Corylus have sporophytic type of self-incompatibility. Several genes related to recognition reaction between pollen and stigma have been identified in hazelnuts. To better understand the self-incompatibility (SI) response, we screened the suitable reference genes by using quantitative real-time reverse transcription PCR (qRT-PCR) analysis in hazelnut for the first time. (2) Methods: the major cultivar "Dawei" was used as material. A total of 12 candidate genes were identified and their expression profiles were compared among different tissues and in response to various treatments (different times after self- and cross-pollination) by RT-qPCR. The expression stability of these 12 candidate reference genes was evaluated using geNorm, NormFinder, BestKeeper, Delta Ct, and RefFinder programs. (3) Results: the comprehensive ranking of RefFinder indicated that ChaActin, VvActin, ChaUBQ14, and ChaEF1-α were the most suitable reference genes. According to the stability analysis of 12 candidate reference genes for each sample group based on four software packages, ChaActin and ChaEF1-α were most stable in different times after self-pollination and 4 h after self- and cross-pollination, respectively. To further validate the suitability of the reference genes identified in this study, CavPrx, which the expression profiles in Corylus have been reported, was quantified by using ChaActin and ChaEF1-α as reference genes. (4) Conclusions: our study of reference genes selection in hazelnut shows that the two reference genes, ChaActin and ChaEF1-α, are suitable for the evaluation of gene expression, and can be used for the analysis of pollen-pistil interaction in Corylus. The results supply a reliable foundation for accurate gene quantifications in Corylus species, which will facilitate the studies related to the reproductive biology in Corylus.
Keywords: Ping’ou hybrid hazelnut (C. heterophylla Fisch. × C. avellana L.); real-time quantitative PCR; reference gene; self-incompatibility; stability of gene expression.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


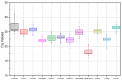
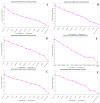


References
-
- Boccacci P., Beltramo C., Sandoval Prando M.A., Lembo A., Sartor C., Mehlenbacher S.A., Torello Marinoni D. In silico mining, characterization and cross-species transferability of EST-SSR markers for European hazelnut (Corylus avellana L.) Mol. Breed. 2015;35:21. doi: 10.1007/s11032-015-0195-7. - DOI
-
- Mehlenbacher S.A. Advances in genetic improvement of hazelnut. Acta Hortic. 2018;1226:12. doi: 10.17660/ActaHortic.2018.1226.1. - DOI
-
- Henry R.J. Forest Trees. Springer; Berlin, Germany: 2011. Wild Crop Relatives: Genomic and Breeding Resources.
-
- Mehlenbacher S.A. Genetic resources for hazelnut: State of the art and future perspectives. Acta Hortic. 2009;845:33–38. doi: 10.17660/ActaHortic.2009.845.1. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

