Double Mutant Cycles as a Tool to Address Folding, Binding, and Allostery
- PMID: 33467625
- PMCID: PMC7830974
- DOI: 10.3390/ijms22020828
Double Mutant Cycles as a Tool to Address Folding, Binding, and Allostery
Abstract
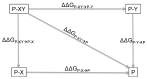
Quantitative measurement of intramolecular and intermolecular interactions in protein structure is an elusive task, not easy to address experimentally. The phenomenon denoted 'energetic coupling' describes short- and long-range interactions between two residues in a protein system. A powerful method to identify and quantitatively characterize long-range interactions and allosteric networks in proteins or protein-ligand complexes is called double-mutant cycles analysis. In this review we describe the thermodynamic principles and basic equations that underlie the double mutant cycle methodology, its fields of application and latest employments, and caveats and pitfalls that the experimentalists must consider. In particular, we show how double mutant cycles can be a powerful tool to investigate allosteric mechanisms in protein binding reactions as well as elusive states in protein folding pathways.
Keywords: coupling energy; interaction networks; site-directed mutagenesis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
Figures

References
-
- Wüthrich K. Protein structure determination in solution by nmr spectroscopy. World Sci. Res. 1995;5:11–14. - PubMed
-
- Fersht A.R. Structure and Mechanism in Protein Science. Freeman; New York, NY, USA: 1999.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

