Evolution and Adaptation of Legionella pneumophila to Manipulate the Ubiquitination Machinery of Its Amoebae and Mammalian Hosts
- PMID: 33467718
- PMCID: PMC7830128
- DOI: 10.3390/biom11010112
Evolution and Adaptation of Legionella pneumophila to Manipulate the Ubiquitination Machinery of Its Amoebae and Mammalian Hosts
Abstract
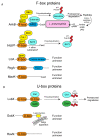
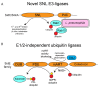
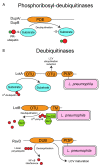
The ubiquitin pathway is highly conserved across the eukaryotic domain of life and plays an essential role in a plethora of cellular processes. It is not surprising that many intracellular bacterial pathogens often target the essential host ubiquitin pathway. The intracellular bacterial pathogen Legionella pneumophila injects into the host cell cytosol multiple classes of classical and novel ubiquitin-modifying enzymes that modulate diverse ubiquitin-related processes in the host cell. Most of these pathogen-injected proteins, designated as effectors, mimic known E3-ubiquitin ligases through harboring F-box or U-box domains. The classical F-box effector, AnkB targets host proteins for K48-linked polyubiquitination, which leads to excessive proteasomal degradation that is required to generate adequate supplies of amino acids for metabolism of the pathogen. In contrast, the SidC and SdcA effectors share no structural similarity to known eukaryotic ligases despite having E3-ubiquitin ligase activity, suggesting that the number of E3-ligases in eukaryotes is under-represented. L. pneumophila also injects into the host many novel ubiquitin-modifying enzymes, which are the SidE family of effectors that catalyze phosphoribosyl-ubiquitination of serine residue of target proteins, independently of the canonical E1-2-3 enzymatic cascade. Interestingly, the environmental bacterium, L. pneumophila, has evolved within a diverse range of amoebal species, which serve as the natural hosts, while accidental transmission through contaminated aerosols can cause pneumonia in humans. Therefore, it is likely that the novel ubiquitin-modifying enzymes of L. pneumophila were acquired by the pathogen through interkingdom gene transfer from the diverse natural amoebal hosts. Furthermore, conservation of the ubiquitin pathway across eukaryotes has enabled these novel ubiquitin-modifying enzymes to function similarly in mammalian cells. Studies on the biological functions of these effectors are likely to reveal further novel ubiquitin biology and shed further lights on the evolution of ubiquitin.
Keywords: Dot/Icm; E1/2-independent ubiquitin ligase; E3-ubiquitin ligase; F-box; SNL E3-ligase; U-box; amoebae; deubiquitinase; effectors; evolution; legionella pneumophila; ubiquitin.
Conflict of interest statement
No potential conflicts of interest were reported by the authors.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

