A SNP in the Cache 1 Signaling Domain of Diguanylate Cyclase STM1987 Leads to Increased In Vivo Fitness of Invasive Salmonella Strains
- PMID: 33468583
- PMCID: PMC8573713
- DOI: 10.1128/IAI.00810-20
A SNP in the Cache 1 Signaling Domain of Diguanylate Cyclase STM1987 Leads to Increased In Vivo Fitness of Invasive Salmonella Strains
Abstract
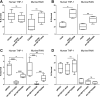
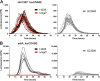
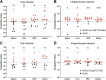
Nontyphoidal Salmonella (NTS) strains are associated with gastroenteritis worldwide but are also the leading cause of bacterial bloodstream infections in sub-Saharan Africa. The invasive NTS (iNTS) strains that cause bloodstream infections differ from standard gastroenteritis-causing strains by >700 single-nucleotide polymorphisms (SNPs). These SNPs are known to alter metabolic pathways and biofilm formation and to contribute to serum resistance and are thought to signify iNTS strains becoming human adapted, similar to typhoid fever-causing Salmonella strains. Identifying SNPs that contribute to invasion or increased virulence has been more elusive. In this study, we identified a SNP in the cache 1 signaling domain of diguanylate cyclase STM1987 in the invasive Salmonella enterica serovar Typhimurium type strain D23580. This SNP was conserved in 118 other iNTS strains analyzed and was comparatively absent in global S Typhimurium isolates associated with gastroenteritis. STM1987 catalyzes the formation of bis-(3',5')-cyclic dimeric GMP (c-di-GMP) and is proposed to stimulate production of cellulose independent of the master biofilm regulator CsgD. We show that the amino acid change in STM1987 leads to a 10-fold drop in cellulose production and increased fitness in a mouse model of acute infection. Reduced cellulose production due to the SNP led to enhanced survival in both murine and human macrophage cell lines. In contrast, loss of CsgD-dependent cellulose production did not lead to any measurable change in in vivo fitness. We hypothesize that the SNP in stm1987 represents a pathoadaptive mutation for iNTS strains.
Keywords: CsgD independent; STM1987; cellulose; cyclic-di-GMP; diguanylate cyclase; invasive Salmonella; macrophage; virulence.
Copyright © 2021 American Society for Microbiology.
Figures


Similar articles
-
Yin and Yang of Biofilm Formation and Cyclic di-GMP Signaling of the Gastrointestinal Pathogen Salmonella enterica Serovar Typhimurium.J Innate Immun. 2022;14(4):275-292. doi: 10.1159/000519573. Epub 2021 Nov 12. J Innate Immun. 2022. PMID: 34775379 Free PMC article. Review.
-
Parallel evolution leading to impaired biofilm formation in invasive Salmonella strains.PLoS Genet. 2019 Jun 24;15(6):e1008233. doi: 10.1371/journal.pgen.1008233. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31233504 Free PMC article.
-
Loss of Multicellular Behavior in Epidemic African Nontyphoidal Salmonella enterica Serovar Typhimurium ST313 Strain D23580.mBio. 2016 Mar 1;7(2):e02265. doi: 10.1128/mBio.02265-15. mBio. 2016. PMID: 26933058 Free PMC article.
-
Cyclic di-GMP signalling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining.Environ Microbiol. 2010 Jan;12(1):40-53. doi: 10.1111/j.1462-2920.2009.02032.x. Epub 2009 Aug 18. Environ Microbiol. 2010. PMID: 19691499
-
Regulation of biofilm formation in Salmonella enterica serovar Typhimurium.Future Microbiol. 2014;9(11):1261-82. doi: 10.2217/fmb.14.88. Future Microbiol. 2014. PMID: 25437188 Review.
Cited by
-
In vitro and ex vivo modeling of enteric bacterial infections.Gut Microbes. 2023 Jan-Dec;15(1):2158034. doi: 10.1080/19490976.2022.2158034. Gut Microbes. 2023. PMID: 36576310 Free PMC article. Review.
-
Genome-wide fitness analysis identifies genes required for in vitro growth and macrophage infection by African and global epidemic pathovariants of Salmonella enterica Enteritidis.Microb Genom. 2023 May;9(5):mgen001017. doi: 10.1099/mgen.0.001017. Microb Genom. 2023. PMID: 37219927 Free PMC article.
-
Yin and Yang of Biofilm Formation and Cyclic di-GMP Signaling of the Gastrointestinal Pathogen Salmonella enterica Serovar Typhimurium.J Innate Immun. 2022;14(4):275-292. doi: 10.1159/000519573. Epub 2021 Nov 12. J Innate Immun. 2022. PMID: 34775379 Free PMC article. Review.
-
Characterisation of Variants of Cyclic di-GMP Turnover Proteins Associated with Semi-Constitutive rdar Morphotype Expression in Commensal and Uropathogenic Escherichia coli Strains.Microorganisms. 2023 Aug 9;11(8):2048. doi: 10.3390/microorganisms11082048. Microorganisms. 2023. PMID: 37630608 Free PMC article.
References
-
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi:10.1371/journal.pmed.1001921. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

