Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
- PMID: 33468660
- PMCID: PMC7848525
- DOI: 10.1073/pnas.2022790118
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Abstract
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is the most common blood disorder, presenting multiple symptoms, including hemolytic anemia. It affects 400 million people worldwide, with more than 160 single mutations reported in G6PD. The most severe mutations (about 70) are classified as class I, leading to more than 90% loss of activity of the wild-type G6PD. The crystal structure of G6PD reveals these mutations are located away from the active site, concentrating around the noncatalytic NADP+-binding site and the dimer interface. However, the molecular mechanisms of class I mutant dysfunction have remained elusive, hindering the development of efficient therapies. To resolve this, we performed integral structural characterization of five G6PD mutants, including four class I mutants, associated with the noncatalytic NADP+ and dimerization, using crystallography, small-angle X-ray scattering (SAXS), cryogenic electron microscopy (cryo-EM), and biophysical analyses. Comparisons with the structure and properties of the wild-type enzyme, together with molecular dynamics simulations, bring forward a universal mechanism for this severe G6PD deficiency due to the class I mutations. We highlight the role of the noncatalytic NADP+-binding site that is crucial for stabilization and ordering two β-strands in the dimer interface, which together communicate these distant structural aberrations to the active site through a network of additional interactions. This understanding elucidates potential paths for drug development targeting G6PD deficiency.
Keywords: G6PD deficiency; NADP+; enzymopathy; hemolytic anemia; structural defects.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

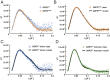


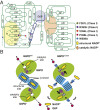
References
-
- Minucci A., et al. , Glucose-6-phosphate dehydrogenase (G6PD) mutations database: Review of the “old” and update of the new mutations. Blood Cells Mol. Dis. 48, 154–165 (2012). - PubMed
-
- Luzzatto L., Nannelli C., Notaro R., Glucose-6-phosphate dehydrogenase deficiency. Hematol. Oncol. Clin. North Am. 30, 373–393 (2016). - PubMed
-
- Nkhoma E. T., Poole C., Vannappagari V., Hall S. A., Beutler E., The global prevalence of glucose-6-phosphate dehydrogenase deficiency: A systematic review and meta-analysis. Blood Cells Mol. Dis. 42, 267–278 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

