Stenoparib, an Inhibitor of Cellular Poly(ADP-Ribose) Polymerase, Blocks Replication of the SARS-CoV-2 and HCoV-NL63 Human Coronaviruses In Vitro
- PMID: 33468703
- PMCID: PMC7845641
- DOI: 10.1128/mBio.03495-20
Stenoparib, an Inhibitor of Cellular Poly(ADP-Ribose) Polymerase, Blocks Replication of the SARS-CoV-2 and HCoV-NL63 Human Coronaviruses In Vitro
Abstract
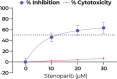
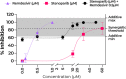
By late 2020, the coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), had caused tens of millions of infections and over 1 million deaths worldwide. A protective vaccine and more effective therapeutics are urgently needed. We evaluated a new poly(ADP-ribose) polymerase (PARP) inhibitor, stenoparib, that recently advanced to phase II clinical trials for treatment of ovarian cancer, for activity against human respiratory coronaviruses, including SARS-CoV-2, in vitro Stenoparib exhibits dose-dependent suppression of SARS-CoV-2 multiplication and spread in Vero E6 monkey kidney and Calu-3 human lung adenocarcinoma cells. Stenoparib was also strongly inhibitory to the human seasonal respiratory coronavirus HCoV-NL63. Compared to remdesivir, which inhibits viral replication downstream of cell entry, stenoparib impedes entry and postentry processes, as determined by time-of-addition (TOA) experiments. Moreover, a 10 μM dosage of stenoparib-below the approximated 25.5 μM half-maximally effective concentration (EC50)-combined with 0.5 μM remdesivir suppressed coronavirus growth by more than 90%, indicating a potentially synergistic effect for this drug combination. Stenoparib as a stand-alone or as part of combinatorial therapy with remdesivir should be a valuable addition to the arsenal against COVID-19.IMPORTANCE New therapeutics are urgently needed in the fight against COVID-19. Repurposing drugs that are either already approved for human use or are in advanced stages of the approval process can facilitate more rapid advances toward this goal. The PARP inhibitor stenoparib may be such a drug, as it is currently in phase II clinical trials for the treatment of ovarian cancer and its safety and dosage in humans have already been established. Our results indicate that stenoparib possesses strong antiviral activity against SARS-CoV-2 and other coronaviruses in vitro. This activity appears to be based on multiple modes of action, where both pre-entry and postentry viral replication processes are impeded. This may provide a therapeutic advantage over many current options that have a narrower target range. Moreover, our results suggest that stenoparib and remdesivir in combination may be especially potent against coronavirus infection.
Keywords: COVID-19; NL63; PARP; SARS-CoV-2; stenoparib.
Copyright © 2021 Stone et al.
Figures




Similar articles
-
Stenoparib, an inhibitor of cellular poly (ADP-ribose) polymerases (PARPs), blocks in vitro replication of SARS-CoV-2 variants.PLoS One. 2022 Sep 14;17(9):e0272916. doi: 10.1371/journal.pone.0272916. eCollection 2022. PLoS One. 2022. PMID: 36103462 Free PMC article.
-
Comparison of Antiviral Activity of Gemcitabine with 2'-Fluoro-2'-Deoxycytidine and Combination Therapy with Remdesivir against SARS-CoV-2.Int J Mol Sci. 2021 Feb 4;22(4):1581. doi: 10.3390/ijms22041581. Int J Mol Sci. 2021. PMID: 33557278 Free PMC article.
-
Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity against Coronaviruses.ACS Infect Dis. 2021 Mar 12;7(3):586-597. doi: 10.1021/acsinfecdis.0c00761. Epub 2021 Mar 1. ACS Infect Dis. 2021. PMID: 33645977 Free PMC article.
-
A Review on Remdesivir: A Possible Promising Agent for the Treatment of COVID-19.Drug Des Devel Ther. 2020 Aug 6;14:3215-3222. doi: 10.2147/DDDT.S261154. eCollection 2020. Drug Des Devel Ther. 2020. PMID: 32821086 Free PMC article. Review.
-
A Review on Remdesivir: A Broad-spectrum Antiviral Molecule for Possible COVID-19 Treatment.Mini Rev Med Chem. 2021;21(17):2530-2543. doi: 10.2174/1389557521666210217093004. Mini Rev Med Chem. 2021. PMID: 33596800 Review.
Cited by
-
A glimpse into viral warfare: decoding the intriguing role of highly pathogenic coronavirus proteins in apoptosis regulation.J Biomed Sci. 2024 Jul 13;31(1):70. doi: 10.1186/s12929-024-01062-1. J Biomed Sci. 2024. PMID: 39003473 Free PMC article. Review.
-
SARS-CoV-2 and the DNA damage response.J Gen Virol. 2023 Nov;104(11):001918. doi: 10.1099/jgv.0.001918. J Gen Virol. 2023. PMID: 37948194 Free PMC article. Review.
-
Novel Drug Design for Treatment of COVID-19: A Systematic Review of Preclinical Studies.Can J Infect Dis Med Microbiol. 2022 Sep 25;2022:2044282. doi: 10.1155/2022/2044282. eCollection 2022. Can J Infect Dis Med Microbiol. 2022. PMID: 36199815 Free PMC article. Review.
-
The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus.J Virol. 2022 Jan 12;96(1):e0096421. doi: 10.1128/JVI.00964-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668775 Free PMC article.
-
NAD+ in COVID-19 and viral infections.Trends Immunol. 2022 Apr;43(4):283-295. doi: 10.1016/j.it.2022.02.001. Epub 2022 Feb 11. Trends Immunol. 2022. PMID: 35221228 Free PMC article. Review.
References
-
- Liu Y, Mao B, Liang S, Yang JW, Lu HW, Chai YH, Wang L, Zhang L, Li QH, Zhao L, He Y, Gu XL, Ji XB, Li L, Jie ZJ, Li Q, Li XY, Lu HZ, Zhang WH, Song YL, Qu JM, Xu JF, Treatment SC. 2020. Association between age and clinical characteristics and outcomes of COVID-19. Eur Respir J 55:2001112. doi:10.1183/13993003.01112-2020. - DOI - PMC - PubMed
-
- Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. 2020. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China (vol 180, pg 934, 2020). JAMA Intern Med 180:934–1031. doi:10.1001/jamainternmed.2020.0994. - DOI - PMC - PubMed
-
- Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen KY, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang JL, Liang Z, Peng Y, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N, Grp CMTE, China Medical Treatment Expert Group for Covid-19. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi:10.1056/NEJMoa2002032. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
