Classification and prediction of protein-protein interaction interface using machine learning algorithm
- PMID: 33469042
- PMCID: PMC7815773
- DOI: 10.1038/s41598-020-80900-2
Classification and prediction of protein-protein interaction interface using machine learning algorithm
Abstract
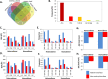
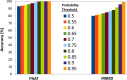
Structural insight of the protein-protein interaction (PPI) interface can provide knowledge about the kinetics, thermodynamics and molecular functions of the complex while elucidating its role in diseases and further enabling it as a potential therapeutic target. However, owing to experimental lag in solving protein-protein complex structures, three-dimensional (3D) knowledge of the PPI interfaces can be gained via computational approaches like molecular docking and post-docking analyses. Despite development of numerous docking tools and techniques, success in identification of native like interfaces based on docking score functions is limited. Hence, we employed an in-depth investigation of the structural features of the interface that might successfully delineate native complexes from non-native ones. We identify interface properties, which show statistically significant difference between native and non-native interfaces belonging to homo and hetero, protein-protein complexes. Utilizing these properties, a support vector machine (SVM) based classification scheme has been implemented to differentiate native and non-native like complexes generated using docking decoys. Benchmarking and comparative analyses suggest very good performance of our SVM classifiers. Further, protein interactions, which are proven via experimental findings but not resolved structurally, were subjected to this approach where 3D-models of the complexes were generated and most likely interfaces were predicted. A web server called Protein Complex Prediction by Interface Properties (PCPIP) is developed to predict whether interface of a given protein-protein dimer complex resembles known protein interfaces. The server is freely available at http://www.hpppi.iicb.res.in/pcpip/ .
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

