Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples
- PMID: 33469065
- PMCID: PMC7815738
- DOI: 10.1038/s41598-020-80352-8
Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples
Abstract
RT-LAMP detection of SARS-CoV-2 has been shown to be a valuable approach to scale up COVID-19 diagnostics and thus contribute to limiting the spread of the disease. Here we present the optimization of highly cost-effective in-house produced enzymes, and we benchmark their performance against commercial alternatives. We explore the compatibility between multiple DNA polymerases with high strand-displacement activity and thermostable reverse transcriptases required for RT-LAMP. We optimize reaction conditions and demonstrate their applicability using both synthetic RNA and clinical patient samples. Finally, we validate the optimized RT-LAMP assay for the detection of SARS-CoV-2 in unextracted heat-inactivated nasopharyngeal samples from 184 patients. We anticipate that optimized and affordable reagents for RT-LAMP will facilitate the expansion of SARS-CoV-2 testing globally, especially in sites and settings where the need for large scale testing cannot be met by commercial alternatives.
Conflict of interest statement
VP was a shareholder at Colorna AB. X. Yin is the co-founder of Biotech & Biomedicine Science (Shenyang) Co. Ltd. S. Ye and X. Liu are employees of Biotech & Biomedicine Science (Shenyang) Co. Ltd. All other authors declare no potential conflict of interest..
Figures


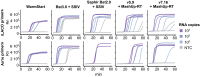

References
-
- Taipale, J., Romer, P. & Linnarsson, S. Population-scale testing can suppress the spread of COVID-19. Preprint at https://www.medrxiv.org/content/10.1101/2020.04.27.20078329v2medRxiv (2020). - DOI
-
- Larremore, D. B. et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. Preprint at https://www.medrxiv.org/content/10.1101/2020.06.22.20136309v3 (2020). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 1580441949000/Natural Science Foundation of Liaoning Province (Liaoning Provincial Natural Science Foundation)/International
- SQ2020YFF0411358/Ministry of Science and Technology of the People's Republic of China (Chinese Ministry of Science and Technology)/International
- KAW 2020.0182/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)/International
- 2016.0123/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)/International
- Swedish Foundations' Starting Grant/Ragnar Söderbergs stiftelse (Ragnar Söderberg Foundation)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

