The Divider Assay is a high-throughput pipeline for aggression analysis in Drosophila
- PMID: 33469118
- PMCID: PMC7815768
- DOI: 10.1038/s42003-020-01617-6
The Divider Assay is a high-throughput pipeline for aggression analysis in Drosophila
Abstract
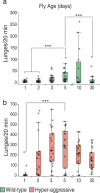
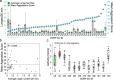
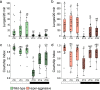
Aggression is a complex social behavior that remains poorly understood. Drosophila has become a powerful model system to study the underlying biology of aggression but lack of high throughput screening and analysis continues to be a barrier for comprehensive mutant and circuit discovery. Here we developed the Divider Assay, a simplified experimental procedure to make aggression analysis in Drosophila fast and accurate. In contrast to existing methods, we can analyze aggression over long time intervals and in complete darkness. While aggression is reduced in the dark, flies are capable of intense fighting without seeing their opponent. Twenty-four-hour behavioral analysis showed a peak in fighting during the middle of the day, a drastic drop at night, followed by re-engagement with a further increase in aggression in anticipation of the next day. Our pipeline is easy to implement and will facilitate high throughput screening for mechanistic dissection of aggression.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Fighting Flies: Quantifying and Analyzing Drosophila Aggression.Cold Spring Harb Protoc. 2023 Sep 1;2023(9):618-627. doi: 10.1101/pdb.top107985. Cold Spring Harb Protoc. 2023. PMID: 37019610
-
Gender-selective patterns of aggressive behavior in Drosophila melanogaster.Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12342-7. doi: 10.1073/pnas.0404693101. Epub 2004 Aug 9. Proc Natl Acad Sci U S A. 2004. PMID: 15302936 Free PMC article.
-
A New Approach that Eliminates Handling for Studying Aggression and the "Loser" Effect in Drosophila melanogaster.J Vis Exp. 2015 Dec 30;(106):e53395. doi: 10.3791/53395. J Vis Exp. 2015. PMID: 26780386 Free PMC article.
-
Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?PLoS Genet. 2015 Aug 27;11(8):e1005416. doi: 10.1371/journal.pgen.1005416. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26312756 Free PMC article. Review.
-
Neuromodulation and Strategic Action Choice in Drosophila Aggression.Annu Rev Neurosci. 2017 Jul 25;40:51-75. doi: 10.1146/annurev-neuro-072116-031240. Epub 2017 Mar 24. Annu Rev Neurosci. 2017. PMID: 28375770 Review.
Cited by
-
Sexual Dimorphism in Aggression: Sex-Specific Fighting Strategies Across Species.Front Behav Neurosci. 2021 Jun 28;15:659615. doi: 10.3389/fnbeh.2021.659615. eCollection 2021. Front Behav Neurosci. 2021. PMID: 34262439 Free PMC article. Review.
-
Drosophila melanogaster as a neurobehavioral model for sex differences in stress response.Front Behav Neurosci. 2025 Jul 11;19:1581763. doi: 10.3389/fnbeh.2025.1581763. eCollection 2025. Front Behav Neurosci. 2025. PMID: 40718064 Free PMC article. Review.
-
Integrating non-mammalian model organisms in the diagnosis of rare genetic diseases in humans.Nat Rev Genet. 2024 Jan;25(1):46-60. doi: 10.1038/s41576-023-00633-6. Epub 2023 Jul 25. Nat Rev Genet. 2024. PMID: 37491400 Review.
-
Drosophila mutants lacking the glial neurotransmitter-modifying enzyme Ebony exhibit low neurotransmitter levels and altered behavior.Sci Rep. 2023 Jun 27;13(1):10411. doi: 10.1038/s41598-023-36558-7. Sci Rep. 2023. PMID: 37369755 Free PMC article.
-
Movement and Dispersion Parameters Characterizing the Group Behavior of Drosophila melanogaster in Micro-Areas of an Observation Arena.Animals (Basel). 2025 May 22;15(11):1515. doi: 10.3390/ani15111515. Animals (Basel). 2025. PMID: 40508981 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

