A genomic view of the microbiome of coral reef demosponges
- PMID: 33469166
- PMCID: PMC8163846
- DOI: 10.1038/s41396-020-00876-9
A genomic view of the microbiome of coral reef demosponges
Abstract
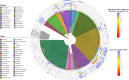
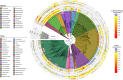
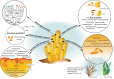
Sponges underpin the productivity of coral reefs, yet few of their microbial symbionts have been functionally characterised. Here we present an analysis of ~1200 metagenome-assembled genomes (MAGs) spanning seven sponge species and 25 microbial phyla. Compared to MAGs derived from reef seawater, sponge-associated MAGs were enriched in glycosyl hydrolases targeting components of sponge tissue, coral mucus and macroalgae, revealing a critical role for sponge symbionts in cycling reef organic matter. Further, visualisation of the distribution of these genes amongst symbiont taxa uncovered functional guilds for reef organic matter degradation. Genes for the utilisation of sialic acids and glycosaminoglycans present in sponge tissue were found in specific microbial lineages that also encoded genes for attachment to sponge-derived fibronectins and cadherins, suggesting these lineages can utilise specific structural elements of sponge tissue. Further, genes encoding CRISPR and restriction-modification systems used in defence against mobile genetic elements were enriched in sponge symbionts, along with eukaryote-like gene motifs thought to be involved in maintaining host association. Finally, we provide evidence that many of these sponge-enriched genes are laterally transferred between microbial taxa, suggesting they confer a selective advantage within the sponge niche and therefore play a critical role in host ecology and evolution.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Knowlton N. Coral reef biodiversity–habitat size matters. Science. 2001;292:1493 LP–1495. - PubMed
-
- Hentschel U, Usher KM, Taylor MW. Marine sponges as microbial fermenters. FEMS Microbiol Ecol. 2006;55:167–77. - PubMed
-
- De Goeij JM, Van Oevelen D, Vermeij MJA, Osinga R, Middelburg JJ, De Goeij AFPM, et al. Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science. 2013;342:108–10. - PubMed
-
- Rix L, de Goeij JM, van Oevelen D, Struck U, Al-Horani FA, Wild C, et al. Differential recycling of coral and algal dissolved organic matter via the sponge loop. Funct Ecol. 2017;31:778–89.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

