Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease
- PMID: 33469207
- PMCID: PMC8035164
- DOI: 10.1038/s41573-020-00108-x
Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease
Abstract
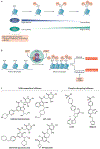
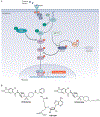
Protein lysine methylation is a crucial post-translational modification that regulates the functions of both histone and non-histone proteins. Deregulation of the enzymes or 'writers' of protein lysine methylation, lysine methyltransferases (KMTs), is implicated in the cause of many diseases, including cancer, mental health disorders and developmental disorders. Over the past decade, significant advances have been made in developing drugs to target KMTs that are involved in histone methylation and epigenetic regulation. The first of these inhibitors, tazemetostat, was recently approved for the treatment of epithelioid sarcoma and follicular lymphoma, and several more are in clinical and preclinical evaluation. Beyond chromatin, the many KMTs that regulate protein synthesis and other fundamental biological processes are emerging as promising new targets for drug development to treat diverse diseases.
Conflict of interest statement
Competing interests
O.G. is a co-founder of EpiCypher and Athelas Therapeutics. J.J. and H.Ü.K. are inventors of patent applications filed by the Icahn School of Medicine at Mount Sinai. J.J. is a consultant of Cullgen and Accent Therapeutics, a scientific advisory board member of Petra Pharma Corporation and an equity shareholder of Cullgen. K.P.B. is an employee at Genentech.
Figures




References
-
- Fischer EH Phosphorylase and the origin of reversible protein phosphorylation. Biol. Chem 391, 131–137 (2010). - PubMed
-
- Wang P, Royer M & Houtz RL Affinity purification of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit epsilon N-methyltransferase. Protein Expr. Purif 6, 528–536 (1995). - PubMed
-
- Rothbart SB & Baylin SB Epigenetic therapy for epithelioid sarcoma. Cell 181, 211 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

