PspA Diversity, Serotype Distribution and Antimicrobial Resistance of Invasive Pneumococcal Isolates from Paediatric Patients in Shenzhen, China
- PMID: 33469319
- PMCID: PMC7810716
- DOI: 10.2147/IDR.S286187
PspA Diversity, Serotype Distribution and Antimicrobial Resistance of Invasive Pneumococcal Isolates from Paediatric Patients in Shenzhen, China
Abstract
Introduction: To determine the phenotypes and genotypes of invasive Streptococcus pneumoniae (S. pneumoniae), 108 strains were isolated from paediatric patients with invasive pneumococcal diseases (IPDs) in Shenzhen from 2014 to 2018.
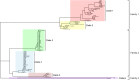
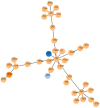
Methods: Serotype profiles were defined by multiplex PCR of the capsule gene. Pneumococcal surface protein A (PspA) classification was performed through pspA gene sequencing. Antimicrobial resistance was examined by broth microdilution. Multilocus sequence typing (MLST) was determined based on next-generation sequencing data.
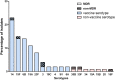
Results: Eighty-one S. pneumoniae of 17 serotypes were finally collected. The coverage of the 13-conjugated polysaccharide vaccine (PCV13) was 88.9%. After the introduction of PCV13, the nonvaccine serotypes were added by serotypes 15b, 16F and 20. Vaccine serotype 3 increased by four serious cases. The pspA family 1 and pspA family 2 are predominant. The multiple drug resistance rate is 91.3%. None of the nonmeningitis isolates were resistant to penicillin, while 98.8% of all the isolates were resistant to erythromycin.
Discussion: This work characterizes the molecular epidemiology of invasive S. pneumoniae in Shenzhen. Continued surveillance of serotype distribution and antimicrobial susceptibility is necessary to alert antibiotic-resistant nonvaccine serotypes and highly virulent serotypes.
Keywords: PspA family; Streptococcus pneumoniae; antimicrobial resistance; invasive pneumococcal disease; serotype.
© 2021 Jiang et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

