Simultaneous Discovery of Positive and Negative Interactions Among Rhizosphere Bacteria Using Microwell Recovery Arrays
- PMID: 33469450
- PMCID: PMC7813777
- DOI: 10.3389/fmicb.2020.601788
Simultaneous Discovery of Positive and Negative Interactions Among Rhizosphere Bacteria Using Microwell Recovery Arrays
Abstract
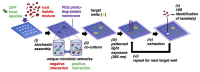
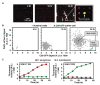
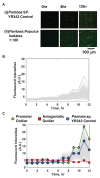
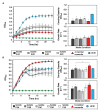
Understanding microbe-microbe interactions is critical to predict microbiome function and to construct communities for desired outcomes. Investigation of these interactions poses a significant challenge due to the lack of suitable experimental tools available. Here we present the microwell recovery array (MRA), a new technology platform that screens interactions across a microbiome to uncover higher-order strain combinations that inhibit or promote the function of a focal species. One experimental trial generates 104 microbial communities that contain the focal species and a distinct random sample of uncharacterized cells from plant rhizosphere. Cells are sequentially recovered from individual wells that display highest or lowest levels of focal species growth using a high-resolution photopolymer extraction system. Interacting species are then identified and putative interactions are validated. Using this approach, we screen the poplar rhizosphere for strains affecting the growth of Pantoea sp. YR343, a plant growth promoting bacteria isolated from Populus deltoides rhizosphere. In one screen, we montiored 3,600 microwells within the array to uncover multiple antagonistic Stenotrophomonas strains and a set of Enterobacter strains that promoted YR343 growth. The later demonstrates the unique ability of the platform to discover multi-membered consortia that generate emergent outcomes, thereby expanding the range of phenotypes that can be characterized from microbiomes. This knowledge will aid in the development of consortia for Populus production, while the platform offers a new approach for screening and discovery of microbial interactions, applicable to any microbiome.
Keywords: consortia; high throughput screening; microbial communities; microbial interactions; microbiome; microdevice; microwell; plant growth promoting rhizobacteria.
Copyright © 2021 Barua, Herken, Stern, Reese, Powers, Morrell-Falvey, Platt and Hansen.
Conflict of interest statement
RH and TP have filed a patent application on this technology. RP and SR were employed by the company Powers & Zhar. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
A Carotenoid-Deficient Mutant in Pantoea sp. YR343, a Bacteria Isolated from the Rhizosphere of Populus deltoides, Is Defective in Root Colonization.Front Microbiol. 2016 Apr 18;7:491. doi: 10.3389/fmicb.2016.00491. eCollection 2016. Front Microbiol. 2016. PMID: 27148182 Free PMC article.
-
Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees.Microbiome. 2017 Feb 23;5(1):25. doi: 10.1186/s40168-017-0241-2. Microbiome. 2017. PMID: 28231859 Free PMC article.
-
Metaproteomics reveals insights into microbial structure, interactions, and dynamic regulation in defined communities as they respond to environmental disturbance.BMC Microbiol. 2021 Nov 8;21(1):308. doi: 10.1186/s12866-021-02370-4. BMC Microbiol. 2021. PMID: 34749649 Free PMC article.
-
Modification of Rhizosphere Microbial Communities: A Possible Mechanism of Plant Growth Promoting Rhizobacteria Enhancing Plant Growth and Fitness.Front Plant Sci. 2022 May 26;13:920813. doi: 10.3389/fpls.2022.920813. eCollection 2022. Front Plant Sci. 2022. PMID: 35720594 Free PMC article. Review.
-
Harnessing microbial multitrophic interactions for rhizosphere microbiome engineering.Microbiol Res. 2022 Dec;265:127199. doi: 10.1016/j.micres.2022.127199. Epub 2022 Sep 15. Microbiol Res. 2022. PMID: 36137486 Review.
Cited by
-
Phenotype testing, genome analysis, and metabolic interactions of three lactic acid bacteria strains existing as a consortium in a naturally fermented milk.Front Microbiol. 2022 Sep 23;13:1000683. doi: 10.3389/fmicb.2022.1000683. eCollection 2022. Front Microbiol. 2022. PMID: 36212860 Free PMC article.
-
Photo-addressable microwell devices for rapid functional screening and isolation of pathogen inhibitors from bacterial strain libraries.Biomicrofluidics. 2024 Feb 29;18(1):014107. doi: 10.1063/5.0188270. eCollection 2024 Jan. Biomicrofluidics. 2024. PMID: 38434239 Free PMC article.
-
Guided by the principles of microbiome engineering: Accomplishments and perspectives for environmental use.mLife. 2022 Nov 3;1(4):382-398. doi: 10.1002/mlf2.12043. eCollection 2022 Dec. mLife. 2022. PMID: 38818482 Free PMC article. Review.
-
Systematic Evaluation of Biotic and Abiotic Factors in Antifungal Microorganism Screening.Microorganisms. 2024 Jul 10;12(7):1396. doi: 10.3390/microorganisms12071396. Microorganisms. 2024. PMID: 39065164 Free PMC article.
-
Low-cost gel-filled microwell array device for screening marine microbial consortium.Front Microbiol. 2022 Dec 16;13:1031439. doi: 10.3389/fmicb.2022.1031439. eCollection 2022. Front Microbiol. 2022. PMID: 36590440 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

