This is a preprint.
Small Molecules to Destabilize the ACE2-RBD Complex: A Molecular Dynamics Study for Potential COVID-19 Therapeutics
- PMID: 33469570
- PMCID: PMC7814830
- DOI: 10.26434/chemrxiv.13377119
Small Molecules to Destabilize the ACE2-RBD Complex: A Molecular Dynamics Study for Potential COVID-19 Therapeutics
Update in
-
Small molecule therapeutics to destabilize the ACE2-RBD complex: A molecular dynamics study.Biophys J. 2021 Jul 20;120(14):2793-2804. doi: 10.1016/j.bpj.2021.06.016. Epub 2021 Jun 30. Biophys J. 2021. PMID: 34214539 Free PMC article.
Abstract
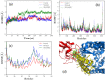
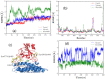
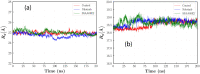
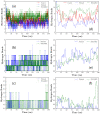
The ongoing COVID-19 pandemic has infected millions of people, claimed hundreds of thousands of lives, and made a worldwide health emergency. Understanding the SARS-CoV-2 mechanism of infection is crucial in the development of potential therapeutics and vaccines. The infection process is triggered by direct binding of the SARS-CoV-2 receptor-binding domain (RBD) to the host cell receptor, Angiotensin-converting enzyme 2 (ACE2). Many efforts have been made to design or repurpose therapeutics to deactivate RBD or ACE2 and prevent the initial binding. In addition to direct inhibition strategies, small chemical compounds might be able to interfere and destabilize the meta-stable, pre-fusion complex of ACE2-RBD. This approach can be employed to prevent the further progress of virus infection at its early stages. In this study, Molecular docking is employed to analyze the binding of two chemical compounds, SSAA09E2 and Nilotinib, with the druggable pocket of the ACE2-RBD complex. The structural changes as a result of the interference with the ACE2-RBD complex are analyzed by molecular dynamics simulations. Results show that both Nilotinib and SSAA09E2 can induce significant conformational changes in the ACE2-RBD complex, intervene with the hydrogen bonds, and influence the flexibility of proteins. Moreover, essential dynamics analysis suggests that the presence of small molecules can trigger large-scale conformational changes that may destabilize the ACE2-RBD complex.
Keywords: ACE2-RBD Complex; COVID-19; Docking; Molecular Dynamics; Nilotinib; SSAA09E2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

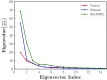
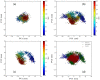

Similar articles
-
Small molecule therapeutics to destabilize the ACE2-RBD complex: A molecular dynamics study.Biophys J. 2021 Jul 20;120(14):2793-2804. doi: 10.1016/j.bpj.2021.06.016. Epub 2021 Jun 30. Biophys J. 2021. PMID: 34214539 Free PMC article.
-
Probing structural basis for enhanced binding of SARS-CoV-2 P.1 variant spike protein with the human ACE2 receptor.J Cell Biochem. 2022 Jul;123(7):1207-1221. doi: 10.1002/jcb.30276. Epub 2022 May 27. J Cell Biochem. 2022. PMID: 35620980 Free PMC article.
-
Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions.mBio. 2021 Mar 30;12(2):e03681-20. doi: 10.1128/mBio.03681-20. mBio. 2021. PMID: 33785634 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
References
-
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., and Wang X., 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. - PubMed
-
- Jahanshahlu L., and Rezaei N., 2020. Monoclonal antibody as a potential anti-COVID-19. /pmc/articles/PMC7269943/?report=abstracthttps://www.ncbi.nlm.nih.gov/pmc/articles/PMC7269943/. - PMC - PubMed
-
- Shanmugaraj B., Siriwattananon K., Wangkanont K., and Phoolcharoen W., 2020. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). https://covid19.elsevierpure.com/en/publications/perspectives-on-monoclo.... - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
