Collateral sensitivity associated with antibiotic resistance plasmids
- PMID: 33470194
- PMCID: PMC7837676
- DOI: 10.7554/eLife.65130
Collateral sensitivity associated with antibiotic resistance plasmids
Abstract
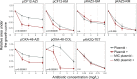
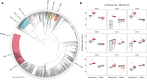
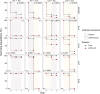
Collateral sensitivity (CS) is a promising alternative approach to counteract the rising problem of antibiotic resistance (ABR). CS occurs when the acquisition of resistance to one antibiotic produces increased susceptibility to a second antibiotic. Recent studies have focused on CS strategies designed against ABR mediated by chromosomal mutations. However, one of the main drivers of ABR in clinically relevant bacteria is the horizontal transfer of ABR genes mediated by plasmids. Here, we report the first analysis of CS associated with the acquisition of complete ABR plasmids, including the clinically important carbapenem-resistance conjugative plasmid pOXA-48. In addition, we describe the conservation of CS in clinical E. coli isolates and its application to selectively kill plasmid-carrying bacteria. Our results provide new insights that establish the basis for developing CS-informed treatment strategies to combat plasmid-mediated ABR.
Keywords: E. coli; antibiotics; collateral sensitivity; evolutionary biology; infectious disease; microbiology; plasmid.
© 2021, Herencias et al.
Conflict of interest statement
CH, JR, RL, AA, JP, RC, ÁS No competing interests declared
Figures


References
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

