Genome-scale de novo assembly using ALGA
- PMID: 33471088
- PMCID: PMC8289375
- DOI: 10.1093/bioinformatics/btab005
Genome-scale de novo assembly using ALGA
Abstract
Motivation: There are very few methods for de novo genome assembly based on the overlap graph approach. It is considered as giving more exact results than the so-called de Bruijn graph approach but in much greater time and of much higher memory usage. It is not uncommon that assembly methods involving the overlap graph model are not able to successfully compute greater datasets, mainly due to memory limitation of a computer. This was the reason for developing in last decades mainly de Bruijn-based assembly methods, fast and fairly accurate. However, the latter methods can fail for longer or more repetitive genomes, as they decompose reads to shorter fragments and lose a part of information. An efficient assembler for processing big datasets and using the overlap graph model is still looked out.
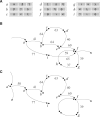
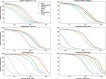
Results: We propose a new genome-scale de novo assembler based on the overlap graph approach, designed for short-read sequencing data. The method, ALGA, incorporates several new ideas resulting in more exact contigs produced in short time. Among these ideas, we have creation of a sparse but quite informative graph, reduction of the graph including a procedure referring to the problem of minimum spanning tree of a local subgraph, and graph traversal connected with simultaneous analysis of contigs stored so far. What is rare in genome assembly, the algorithm is almost parameter-free, with only one optional parameter to be set by a user. ALGA was compared with nine state-of-the-art assemblers in tests on genome-scale sequencing data obtained from real experiments on six organisms, differing in size, coverage, GC content and repetition rate. ALGA produced best results in the sense of overall quality of genome reconstruction, understood as a good balance between genome coverage, accuracy and length of resulting sequences. The algorithm is one of tools involved in processing data in currently realized national project Genomic Map of Poland.
Availability and implementation: ALGA is available at http://alga.put.poznan.pl.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press.
Figures

References
-
- Ameur A. et al. (2019) Single-molecule sequencing: towards clinical applications. Trends Biotechnol., 37, 72–85. - PubMed
-
- Blazewicz J. et al. (2009) Whole genome assembly from 454 sequencing output via modified DNA graph concept. Comput. Biol. Chem., 33, 224–230. - PubMed
-
- Blazewicz J. et al. (2002) A heuristic managing errors for DNA sequencing. Bioinformatics, 18, 652–660. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

