Therapeutic targeting of transcriptional elongation in diffuse intrinsic pontine glioma
- PMID: 33471107
- PMCID: PMC8328031
- DOI: 10.1093/neuonc/noab009
Therapeutic targeting of transcriptional elongation in diffuse intrinsic pontine glioma
Abstract
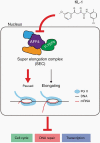
Background: Diffuse intrinsic pontine glioma (DIPG) is associated with transcriptional dysregulation driven by H3K27 mutation. The super elongation complex (SEC) is required for transcriptional elongation through release of RNA polymerase II (Pol II). Inhibition of transcription elongation by SEC disruption can be an effective therapeutic strategy of H3K27M-mutant DIPG. Here, we tested the effect of pharmacological disruption of the SEC in H3K27M-mutant DIPG to advance understanding of the molecular mechanism and as a new therapeutic strategy for DIPG.
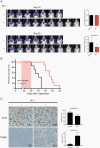
Methods: Short hairpin RNAs (shRNAs) were used to suppress the expression of AF4/FMR2 4 (AFF4), a central SEC component, in H3K27M-mutant DIPG cells. A peptidomimetic lead compound KL-1 was used to disrupt a functional component of SEC. Cell viability assay, colony formation assay, and apoptosis assay were utilized to analyze the effects of KL-1 treatment. RNA- and ChIP-sequencing were used to determine the effects of KL-1 on gene expression and chromatin occupancy. We treated mice bearing H3K27M-mutant DIPG patient-derived xenografts (PDXs) with KL-1. Intracranial tumor growth was monitored by bioluminescence image and therapeutic response was evaluated by animal survival.
Results: Depletion of AFF4 significantly reduced the cell growth of H3K27M-mutant DIPG. KL-1 increased genome-wide Pol II occupancy and suppressed transcription involving multiple cellular processes that promote cell proliferation and differentiation of DIPG. KL-1 treatment suppressed DIPG cell growth, increased apoptosis, and prolonged animal survival with H3K27M-mutant DIPG PDXs.
Conclusions: SEC disruption by KL-1 increased therapeutic benefit in vitro and in vivo, supporting a potential therapeutic activity of KL-1 in H3K27M-mutant DIPG.
Keywords: H3K27M-mutant DIPG; RNA polymerase II (Pol II); patient-derived xenograft (PDX); super elongation complex (SEC); transcriptional elongation.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



Comment in
-
Converging evidence for inhibition of transcriptional control in high-grade gliomas.Neuro Oncol. 2021 Aug 2;23(8):1225-1227. doi: 10.1093/neuonc/noab112. Neuro Oncol. 2021. PMID: 33984150 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

