Covid-19 Automated Diagnosis and Risk Assessment through Metabolomics and Machine Learning
- PMID: 33471512
- PMCID: PMC8023531
- DOI: 10.1021/acs.analchem.0c04497
Covid-19 Automated Diagnosis and Risk Assessment through Metabolomics and Machine Learning
Abstract
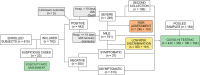
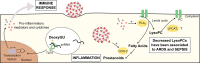
COVID-19 is still placing a heavy health and financial burden worldwide. Impairment in patient screening and risk management plays a fundamental role on how governments and authorities are directing resources, planning reopening, as well as sanitary countermeasures, especially in regions where poverty is a major component in the equation. An efficient diagnostic method must be highly accurate, while having a cost-effective profile. We combined a machine learning-based algorithm with mass spectrometry to create an expeditious platform that discriminate COVID-19 in plasma samples within minutes, while also providing tools for risk assessment, to assist healthcare professionals in patient management and decision-making. A cross-sectional study enrolled 815 patients (442 COVID-19, 350 controls and 23 COVID-19 suspicious) from three Brazilian epicenters from April to July 2020. We were able to elect and identify 19 molecules related to the disease's pathophysiology and several discriminating features to patient's health-related outcomes. The method applied for COVID-19 diagnosis showed specificity >96% and sensitivity >83%, and specificity >80% and sensitivity >85% during risk assessment, both from blinded data. Our method introduced a new approach for COVID-19 screening, providing the indirect detection of infection through metabolites and contextualizing the findings with the disease's pathophysiology. The pairwise analysis of biomarkers brought robustness to the model developed using machine learning algorithms, transforming this screening approach in a tool with great potential for real-world application.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Döhla M.; Boesecke C.; Schulte B.; Diegmann C.; Sib E.; Richter E.; Eschbach-Bludau M.; Aldabbagh S.; Marx B.; Eis-Hübinger A.-M. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health 2020, 182, 170–172. 10.1016/j.puhe.2020.04.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

