Genetic Evaluation of Nosocomial Candida auris Transmission
- PMID: 33472901
- PMCID: PMC8092739
- DOI: 10.1128/JCM.02252-20
Genetic Evaluation of Nosocomial Candida auris Transmission
Abstract
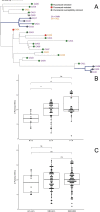
Whole-genome sequences of Candida auris isolates from nosocomial and nonnosocomial infections were compared. The average numbers of single nucleotide variations were different between the two groups. The small amount of genetic variability between intra- or interhost isolates suggests recovery of all colonizing or infecting genomes for comparison is required for outbreaks.
Keywords: Candida auris; nosocomial transmission; whole-genome sequencing.
Copyright © 2021 American Society for Microbiology.
Figures
References
-
- CDC. 2020. CDC & FDA Antibiotic Resistance Isolate Bank. https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=2. Accessed 14 May 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

